Transition-transversion bias is not universal: a counter example from grasshopper pseudogenes
- PMID: 17274688
- PMCID: PMC1790724
- DOI: 10.1371/journal.pgen.0030022
Transition-transversion bias is not universal: a counter example from grasshopper pseudogenes
Abstract
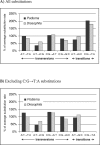
Comparisons of the DNA sequences of metazoa show an excess of transitional over transversional substitutions. Part of this bias is due to the relatively high rate of mutation of methylated cytosines to thymine. Postmutation processes also introduce a bias, particularly selection for codon-usage bias in coding regions. It is generally assumed, however, that there is a universal bias in favour of transitions over transversions, possibly as a result of the underlying chemistry of mutation. Surprisingly, this underlying trend has been evaluated only in two types of metazoan, namely Drosophila and the Mammalia. Here, we investigate a third group, and find no such bias. We characterize the point substitution spectrum in Podisma pedestris, a grasshopper species with a very large genome. The accumulation of mutations was surveyed in two pseudogene families, nuclear mitochondrial and ribosomal DNA sequences. The cytosine-guanine (CpG) dinucleotides exhibit the high transition frequencies expected of methylated sites. The transition rate at other cytosine residues is significantly lower. After accounting for this methylation effect, there is no significant difference between transition and transversion rates. These results contrast with reports from other taxa and lead us to reject the hypothesis of a universal transition/transversion bias. Instead we suggest fundamental interspecific differences in point substitution processes.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


References
-
- Graur D, Li WH. Fundamentals of molecular evolution. Sunderland, MA: Sinauer Associates; 2000. 481
-
- Zhang F, Zhao Z. The influence of neighboring-nucleotide composition on single nucleotide polymorphisms (SNPs) in the mouse genome and its comparison with human SNPs. Genomics. 2004;84:785–795. - PubMed
-
- Bensasson D, Petrov DA, Zhang D-X, Hartl DL, Hewitt GM. Genomic gigantism: DNA loss is slow in mountain grasshoppers. Mol Biol Evol. 2001;18:246–253. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

