Transcriptional pathways in second heart field development
- PMID: 17276708
- PMCID: PMC1855211
- DOI: 10.1016/j.semcdb.2007.01.001
Transcriptional pathways in second heart field development
Abstract
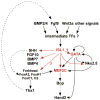
The heart is the first organ to form and function during vertebrate development and is absolutely essential for life. The left ventricle is derived from the classical primary or first heart field (FHF), while the right ventricle and outflow tract are derived from a distinct second heart field (SHF). The recent discovery of the SHF has raised several fundamental and important questions about how the two heart fields are integrated into a single organ and whether unique molecular programs control the development of the two heart fields. This review briefly highlights the contributions of the SHF to the developing and mature heart and then focuses primarily on our current understanding of the transcriptional pathways that function in the development of the SHF and its derivatives.
Figures


References
-
- Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. - PubMed
-
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–48. - PubMed
-
- Srivastava D. Genetic assembly of the heart: implications for congenital heart disease. Annu Rev Physiol. 2001;63:451–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

