SCOOP: a simple method for identification of novel protein superfamily relationships
- PMID: 17277330
- PMCID: PMC2603044
- DOI: 10.1093/bioinformatics/btm034
SCOOP: a simple method for identification of novel protein superfamily relationships
Abstract
Motivation: Profile searches of sequence databases are a sensitive way to detect sequence relationships. Sophisticated profile-profile comparison algorithms that have been recently introduced increase search sensitivity even further.
Results: In this article, a simpler approach than profile-profile comparison is presented that has a comparable performance to state-of-the-art tools such as COMPASS, HHsearch and PRC. This approach is called SCOOP (Simple Comparison Of Outputs Program), and is shown to find known relationships between families in the Pfam database as well as detect novel distant relationships between families. Several novel discoveries are presented including the discovery that a domain of unknown function (DUF283) found in Dicer proteins is related to double-stranded RNA-binding domains.
Availability: SCOOP is freely available under a GNU GPL license from http://www.sanger.ac.uk/Users/agb/SCOOP/.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
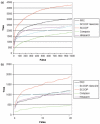

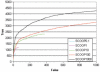



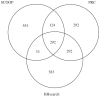
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

