Power and precision of alternate methods for linkage disequilibrium mapping of quantitative trait loci
- PMID: 17277369
- PMCID: PMC1855130
- DOI: 10.1534/genetics.106.066480
Power and precision of alternate methods for linkage disequilibrium mapping of quantitative trait loci
Abstract
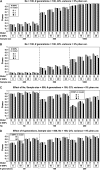
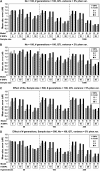
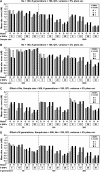
Linkage disequilibrium (LD) analysis in outbred populations uses historical recombinations to detect and fine map quantitative trait loci (QTL). Our objective was to evaluate the effect of various factors on power and precision of QTL detection and to compare LD mapping methods on the basis of regression and identity by descent (IBD) in populations of limited effective population size (N(e)). An 11-cM region with 6-38 segregating single-nucleotide polymorphisms (SNPs) and a central QTL was simulated. After 100 generations of random mating with N(e) of 50, 100, or 200, SNP genotypes and phenotypes were generated on 200, 500, or 1000 individuals with the QTL explaining 2 or 5% of phenotypic variance. To detect and map the QTL, phenotypes were regressed on genotypes or (assumed known) haplotypes, in comparison with the IBD method. Power and precision to detect QTL increased with sample size, marker density, and QTL effect. Power decreased with N(e), but precision was affected little by N(e). Single-marker regression had similar or greater power and precision than other regression models, and was comparable to the IBD method. Thus, for rapid initial screening of samples of adequate size in populations in which drift is the primary force that has created LD, QTL can be detected and mapped by regression on SNP genotypes without recovering haplotypes.
Figures
Similar articles
-
Optimal haplotype structure for linkage disequilibrium-based fine mapping of quantitative trait loci using identity by descent.Genetics. 2006 Mar;172(3):1955-65. doi: 10.1534/genetics.105.048686. Epub 2005 Dec 1. Genetics. 2006. PMID: 16322505 Free PMC article.
-
Comparing linkage disequilibrium-based methods for fine mapping quantitative trait loci.Genetics. 2004 Mar;166(3):1561-70. doi: 10.1534/genetics.166.3.1561. Genetics. 2004. PMID: 15082569 Free PMC article.
-
Multipoint identity-by-descent prediction using dense markers to map quantitative trait loci and estimate effective population size.Genetics. 2007 Aug;176(4):2551-60. doi: 10.1534/genetics.107.070953. Epub 2007 Jun 11. Genetics. 2007. PMID: 17565953 Free PMC article.
-
QTL mapping using high-throughput sequencing.Methods Mol Biol. 2015;1284:257-85. doi: 10.1007/978-1-4939-2444-8_13. Methods Mol Biol. 2015. PMID: 25757777 Review.
-
Methods for linkage disequilibrium mapping in crops.Trends Plant Sci. 2007 Feb;12(2):57-63. doi: 10.1016/j.tplants.2006.12.001. Epub 2007 Jan 16. Trends Plant Sci. 2007. PMID: 17224302 Review.
Cited by
-
Genotype matrix mapping: searching for quantitative trait loci interactions in genetic variation in complex traits.DNA Res. 2007 Oct 31;14(5):217-25. doi: 10.1093/dnares/dsm020. Epub 2007 Nov 13. DNA Res. 2007. PMID: 18000014 Free PMC article.
-
Single Marker and Haplotype-Based Association Analysis of Semolina and Pasta Colour in Elite Durum Wheat Breeding Lines Using a High-Density Consensus Map.PLoS One. 2017 Jan 30;12(1):e0170941. doi: 10.1371/journal.pone.0170941. eCollection 2017. PLoS One. 2017. PMID: 28135299 Free PMC article.
-
Genome wide association studies for milk production traits in Chinese Holstein population.PLoS One. 2010 Oct 27;5(10):e13661. doi: 10.1371/journal.pone.0013661. PLoS One. 2010. PMID: 21048968 Free PMC article.
-
A Novel QTL for Resistance to Phytophthora Crown Rot in Squash.Plants (Basel). 2021 Oct 6;10(10):2115. doi: 10.3390/plants10102115. Plants (Basel). 2021. PMID: 34685924 Free PMC article.
-
Fine-mapping quantitative trait loci with a medium density marker panel: efficiency of population structures and comparison of linkage disequilibrium linkage analysis models.Genet Res (Camb). 2012 Aug;94(4):223-34. doi: 10.1017/S0016672312000407. Genet Res (Camb). 2012. PMID: 22950902 Free PMC article.
References
-
- Bonnen, P. E., I. Pe'er, R. M. Plenge, J. Salit, J. K. Lowe et al., 2006. Evaluating potential for whole-genome studies in Kosrae, an isolated population in Micronesia. Nat. Genet. 38: 214–217. - PubMed
-
- Dekkers, J. C. M., and F. Hospital, 2002. The use of molecular genetics in the improvement of agricultural populations. Nat. Rev. Genet. 3: 22–32. - PubMed
-
- Eller, E., J. Hawks and J. H. Relethford, 2004. Local extinction and recolonization, species effective population size, and modern human origins. Hum. Biol. 76: 689–709. - PubMed
-
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics. Addison-Wesley Longman, Harlow, UK.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

