Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris
- PMID: 17277373
- PMCID: PMC1855124
- DOI: 10.1534/genetics.106.064469
Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris
Abstract
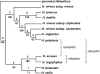
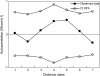
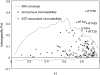
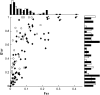
Plant species may remain morphologically distinct despite gene exchange with congeners, yet little is known about the genomewide pattern of introgression among species. Here we analyze the effects of persistent gene flow on genomic differentiation between the sympatric sunflower species Helianthus annuus and H. petiolaris. While the species are strongly isolated in testcrosses, genetic distances at 108 microsatellite loci and 14 sequenced genes are highly variable and much lower (on average) than for more closely related but historically allopatric congeners. Our analyses failed to detect a positive association between levels of genetic differentiation and chromosomal rearrangements (as reported in a prior publication) or proximity to QTL for morphological differences or hybrid sterility. However, a significant increase in differentiation was observed for markers within 5 cM of chromosomal breakpoints. Together, these results suggest that islands of differentiation between these two species are small, except in areas of low recombination. Furthermore, only microsatellites associated with ESTs were identified as outlier loci in tests for selection, which might indicate that the ESTs themselves are the targets of selection rather than linked genes (or that coding regions are not randomly distributed). In general, these results indicate that even strong and genetically complex reproductive barriers cannot prevent widespread introgression.
Figures


References
-
- Arnold, M. L., 1997. Natural Hybridization and Evolution. Oxford University Press, New York.
-
- Barton, N. H., 1979. Dynamics of hybrid zones. Heredity 43: 341–359.
-
- Barton, N. H., and G. M. Hewitt, 1985. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 16: 113–148.
-
- Beaumont, M. A., and R. A. Nichols, 1996. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B Biol. Sci. 263: 1619–1626.
-
- Buerkle, C. A., and L. H. Rieseberg, 2001. Low intraspecific variation for genomic isolation between hybridizing sunflower species. Evolution 55: 684–691. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

