Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice
- PMID: 17283043
- PMCID: PMC1899923
- DOI: 10.1128/MCB.01652-06
Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice
Abstract
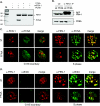
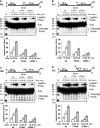
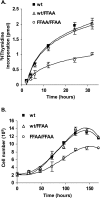
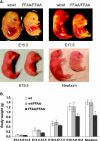
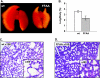
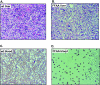
The interaction between flap endonuclease 1 (FEN-1) and proliferation cell nuclear antigen (PCNA) is critical for faithful and efficient Okazaki fragment maturation. In a living cell, this interaction is probably important for PCNA to load FEN-1 to the replication fork, to coordinate the sequential functions of FEN-1 and other enzymes, and to stimulate its enzyme activity. The FEN-1/PCNA interaction is mediated by the motif (337)QGRLDDFFK(345) of FEN-1, such that an F343AF344A (FFAA) mutant cannot bind to PCNA but retains its nuclease activities. To determine the physiological roles of the FEN-1/PCNA interaction in a mammalian system, we knocked the FFAA Fen1 mutation into the Fen1 gene locus of mice. FFAA/FFAA mouse embryo fibroblasts underwent DNA replication and division at a slower pace, and FFAA/FFAA mutant embryos displayed significant defects in growth and development, particularly in the lung and blood systems. All newborn FFAA mutant pups died at birth, likely due to pulmonary hypoplasia and pancytopenia. Collectively, our data demonstrate the importance of the FEN-1/PCNA complex in DNA replication and in the embryonic development of mice.
Figures

References
-
- Adachi, N., Z. E. Karanjawala, Y. Matsuzaki, H. Koyama, and M. R. Lieber. 2002. Two overlapping divergent transcription units in the human genome: the FEN1/C11orf10 locus. OMICS J. Integr. Biol. 6:273-279. - PubMed
-
- Ayyagari, R., X. V. Gomes, D. A. Gordenin, and P. M. Burgers. 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278:1618-1625. - PubMed
-
- Bae, S. H., K. H. Bae, J. A. Kim, and Y. S. Seo. 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412:456-461. - PubMed
-
- Biswas, E. E., F. X. Zhu, and S. B. Biswas. 1997. Stimulation of RTH1 nuclease of the yeast Saccharomyces cerevisiae by replication protein A. Biochemistry 36:5955-5962. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
