A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling
- PMID: 17283044
- PMCID: PMC1899937
- DOI: 10.1128/MCB.02323-06
A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling
Abstract
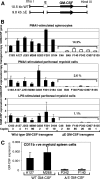
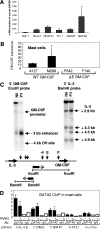
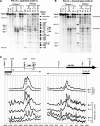
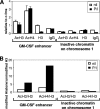
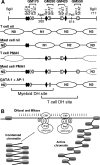
We investigated alternate mechanisms employed by enhancers to position and remodel nucleosomes and activate tissue-specific genes in divergent cell types. We demonstrated that the granulocyte-macrophage colony-stimulating factor (GM-CSF) gene enhancer is modular and recruits different sets of transcription factors in T cells and myeloid cells. The enhancer recruited distinct inducible tissue-specific enhanceosome-like complexes and directed nucleosomes to different positions in these cell types. In undifferentiated T cells, the enhancer was activated by inducible binding of two NFAT/AP-1 complexes which disrupted two specifically positioned nucleosomes (N1 and N2). In myeloid cells, the enhancer was remodeled by GATA factors which constitutively displaced an upstream nucleosome (N0) and cooperated with inducible AP-1 elements to activate transcription. In mast cells, which express both GATA-2 and NFAT, these two pathways combined to activate the enhancer and generate high-level gene expression. At least 5 kb of the GM-CSF locus was organized as an array of nucleosomes with fixed positions, but the enhancer adopted different nucleosome positions in T cells and mast cells. Furthermore, nucleosomes located between the enhancer and promoter were mobilized upon activation in an enhancer-dependent manner. These studies reveal that distinct tissue-specific mechanisms can be used either alternately or in combination to activate the same enhancer.
Figures




References
-
- Avots, A., M. Buttmann, S. Chuvpilo, C. Escher, U. Smola, A. J. Bannister, U. R. Rapp, T. Kouzarides, and E. Serfling. 1999. CBP/p300 integrates Raf/Rac-signaling pathways in the transcriptional induction of NF-ATc during T cell activation. Immunity 10:515-524. - PubMed
-
- Bert, A. G., J. Burrows, A. Hawwari, M. A. Vadas, and P. N. Cockerill. 2000. Reconstitution of T cell-specific transcription directed by composite NFAT/Oct elements. J. Immunol. 165:5646-5655. - PubMed
-
- Butterfield, J. H., and D. A. Weiler. 1989. In vitro sensitivity of immature human mast cells to chemotherapeutic agents. Int. Arch. Allergy Appl. Immunol. 89:297-300. - PubMed
-
- Carey, M. 1998. The enhanceosome and transcriptional synergy. Cell 92:5-8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
