Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation
- PMID: 17283045
- PMCID: PMC1899951
- DOI: 10.1128/MCB.01339-06
Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation
Abstract
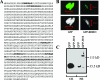
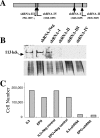
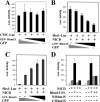
RBM15 is the fusion partner with MKL in the t(1;22) translocation of acute megakaryoblastic leukemia. To understand the role of the RBM15-MKL1 fusion protein in leukemia, we must understand the normal functions of RBM15 and MKL. Here, we show a role for Rbm15 in myelopoiesis. Rbm15 is expressed at highest levels in hematopoietic stem cells and at more moderate levels during myelopoiesis of murine cell lines and primary murine cells. Decreasing Rbm15 levels with RNA interference enhances differentiation of the 32DWT18 myeloid precursor cell line. Conversely, enforced expression of Rbm15 inhibits 32DWT18 differentiation. We show that Rbm15 alters Notch-induced HES1 promoter activity in a cell type-specific manner. Rbm15 inhibits Notch-induced HES1 transcription in nonhematopoietic cells but stimulates this activity in hematopoietic cell lines, including 32DWT18 and human erythroleukemia cells. Moreover, the N terminus of Rbm15 coimmunoprecipitates with RBPJkappa, a critical factor in Notch signaling, and the Rbm15 N terminus has a dominant negative effect, impairing activation of HES1 promoter activity by full-length-Rbm15. Thus, Rbm15 is differentially expressed during hematopoiesis and may act to inhibit myeloid differentiation in hematopoietic cells via a mechanism that is mediated by stimulation of Notch signaling via RBPJkappa.
Figures



References
-
- Ballerini, P., A. Blaise, T. Mercher, B. Pellegrino, C. Perot, J. van den Akker, E. Gatbois, M. Adam, L. Douay, R. Berger, O. Bernard, and J. Landman-Parker. 2003. A novel real-time RT-PCR assay for quantification of OTT-MAL fusion transcript reliable for diagnosis of t(1;22) and minimal residual disease (MRD) detection. Leukemia 17:1193-1196. - PubMed
-
- Baruchel, A., M. T. Daniel, G. Schaison, and R. Berger. 1991. Nonrandom t(1;22)(p12-p13;q13) in acute megakaryocytic malignant proliferation. Cancer Genet. Cytogenet. 54:239-243. - PubMed
-
- Beatus, P., J. Lundkvist, C. Oberg, and U. Lendahl. 1999. The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development 126:3925-3935. - PubMed
-
- Carroll, A., C. Civin, N. Schneider, G. Dahl, A. Pappo, P. Bowman, A. Emami, S. Gross, C. Alvarado, C. Phillips, et al. 1991. The t(1;22) (p13;q13) is nonrandom and restricted to infants with acute megakaryoblastic leukemia. Blood 78:748-752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
