Bmp2 transcription in osteoblast progenitors is regulated by a distant 3' enhancer located 156.3 kilobases from the promoter
- PMID: 17283059
- PMCID: PMC1899916
- DOI: 10.1128/MCB.01609-06
Bmp2 transcription in osteoblast progenitors is regulated by a distant 3' enhancer located 156.3 kilobases from the promoter
Abstract
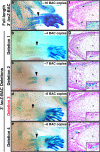
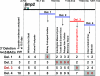
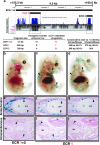
Bone morphogenetic protein 2 (encoded by Bmp2) has been implicated as an important signaling ligand for osteoblast differentiation and bone formation and as a genetic risk factor for osteoporosis. To initially survey a large genomic region flanking the mouse Bmp2 gene for cis-regulatory function, two bacterial artificial chromosome (BAC) clones that extend far upstream and downstream of the gene were engineered to contain a lacZ reporter cassette and tested in transgenic mice. Each BAC clone directs a distinct subset of normal Bmp2 expression patterns, suggesting a modular arrangement of distant Bmp2 regulatory elements. Strikingly, regulatory sequences required for Bmp2 expression in differentiating osteoblasts, as well as tooth buds, hair placodes, kidney, and other tissues, are located more than 53 kilobases 3' to the promoter. By testing BACs with engineered deletions across this distant 3' region, we parsed these regulatory elements into separate locations and more closely refined the location of the osteoblast progenitor element. Finally, a conserved osteoblast progenitor enhancer was identified within a 656-bp sequence located 156.3 kilobases 3' from the promoter. The identification of this enhancer should permit further investigation of upstream regulatory mechanisms that control Bmp2 transcription during osteoblast differentiation and are relevant to further studies of Bmp2 as a candidate risk factor gene for osteoporosis.
Figures






References
-
- Abrams, K. L., J. Xu, C. Nativelle-Serpentini, S. Dabirshahsahebi, and M. B. Rogers. 2004. An evolutionary and molecular analysis of Bmp2 expression. J. Biol. Chem. 279:15916-15928. - PubMed
-
- Aubin, J. E. 1998. Advances in the osteoblast lineage. Biochem. Cell Biol. 76:899-910. - PubMed
-
- Berman, B. P., Y. Nibu, B. D. Pfeiffer, P. Tomancak, S. E. Celniker, M. Levine, G. M. Rubin, and M. B. Eisen. 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99:757-762. - PMC - PubMed
-
- Bitgood, M. J., and A. P. McMahon. 1995. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev. Biol. 172:126-138. - PubMed
-
- Blackman, R. K., M. Sanicola, L. A. Raftery, T. Gillevet, and W. M. Gelbart. 1991. An extensive 3′ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development 111:657-666. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
