Dependency of ISW1a chromatin remodeling on extranucleosomal DNA
- PMID: 17283061
- PMCID: PMC1899934
- DOI: 10.1128/MCB.01731-06
Dependency of ISW1a chromatin remodeling on extranucleosomal DNA
Abstract
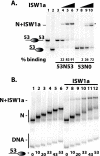
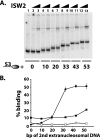
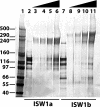
The nucleosome remodeling activity of ISW1a was dependent on whether ISW1a was bound to one or both extranucleosomal DNAs. ISW1a preferentially bound nucleosomes with an optimal length of approximately 33 to 35 bp of extranucleosomal DNA at both the entry and exit sites over nucleosomes with extranucleosomal DNA at only one entry or exit site. Nucleosomes with extranucleosomal DNA at one of the entry/exit sites were readily remodeled by ISW1a and stimulated the ATPase activity of ISW1a, while conversely, nucleosomes with extranucleosomal DNA at both entry/exit sites were unable either to stimulate the ATPase activity of ISW1a or to be mobilized. DNA footprinting revealed that a major conformational difference between the nucleosomes was the lack of ISW1a binding to nucleosomal DNA two helical turns from the dyad axis in nucleosomes with extranucleosomal DNA at both entry/exit sites. The Ioc3 subunit of ISW1a was found to be the predominant subunit associated with extranucleosomal DNA when ISW1a is bound either to one or to both extranucleosomal DNAs. These two conformations of the ISW1a-nucleosome complex are suggested to be the molecular basis for the nucleosome spacing activity of ISW1a on nucleosomal arrays. ISW1b, the other isoform of ISW1, does not have the same dependency for extranucleosomal DNA as ISW1a and, likewise, is not able to space nucleosomes.
Figures




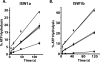
Similar articles
-
Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2.Mol Cell Biol. 2004 Nov;24(22):10047-57. doi: 10.1128/MCB.24.22.10047-10057.2004. Mol Cell Biol. 2004. PMID: 15509805 Free PMC article.
-
Isw1a does not have strict limitations on the length of extranucleosomal DNAs for mobilization of nucleosomes assembled with HeLa cell histones.J Biomol Struct Dyn. 2014 Apr;32(4):523-31. doi: 10.1080/07391102.2013.782823. Epub 2013 Apr 13. J Biomol Struct Dyn. 2014. PMID: 23581908
-
Yeast Isw1a and Isw1b exhibit similar nucleosome mobilization capacities for mononucleosomes, but differently mobilize dinucleosome templates.Arch Biochem Biophys. 2014 Mar 15;546:72-80. doi: 10.1016/j.abb.2014.02.003. Epub 2014 Feb 14. Arch Biochem Biophys. 2014. PMID: 24530316
-
Comparison of the Isw1a, Isw1b, and Isw2 nucleosome disrupting activities.Biochemistry. 2013 Oct 8;52(40):6940-9. doi: 10.1021/bi400634r. Epub 2013 Sep 27. Biochemistry. 2013. PMID: 24050724
-
Remodeling chromatin structures for transcription: what happens to the histones?Bioessays. 1996 Nov;18(11):875-84. doi: 10.1002/bies.950181106. Bioessays. 1996. PMID: 8939065 Review.
Cited by
-
Structure of the ISW1a complex bound to the dinucleosome.Nat Struct Mol Biol. 2024 Feb;31(2):266-274. doi: 10.1038/s41594-023-01174-6. Epub 2024 Jan 4. Nat Struct Mol Biol. 2024. PMID: 38177688
-
ACF catalyses chromatosome movements in chromatin fibres.EMBO J. 2008 Mar 19;27(6):817-26. doi: 10.1038/sj.emboj.7601902. Epub 2007 Oct 25. EMBO J. 2008. PMID: 17962805 Free PMC article.
-
Ruler elements in chromatin remodelers set nucleosome array spacing and phasing.Nat Commun. 2021 May 28;12(1):3232. doi: 10.1038/s41467-021-23015-0. Nat Commun. 2021. PMID: 34050140 Free PMC article.
-
Genomic Nucleosome Organization Reconstituted with Pure Proteins.Cell. 2016 Oct 20;167(3):709-721.e12. doi: 10.1016/j.cell.2016.09.045. Cell. 2016. PMID: 27768892 Free PMC article.
-
Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders.Proc Natl Acad Sci U S A. 2012 Nov 20;109(47):19238-43. doi: 10.1073/pnas.1213825109. Epub 2012 Nov 7. Proc Natl Acad Sci U S A. 2012. PMID: 23134727 Free PMC article.
References
-
- Boyer, L. A., M. R. Langer, K. A. Crowley, S. Tan, J. M. Denu, and C. L. Peterson. 2002. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10:935-942. - PubMed
-
- Boyer, L. A., R. R. Latek, and C. L. Peterson. 2004. The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5:158-163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
