Use of a cAMP BRET sensor to characterize a novel regulation of cAMP by the sphingosine 1-phosphate/G13 pathway
- PMID: 17283075
- PMCID: PMC2526465
- DOI: 10.1074/jbc.M609695200
Use of a cAMP BRET sensor to characterize a novel regulation of cAMP by the sphingosine 1-phosphate/G13 pathway
Abstract
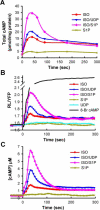
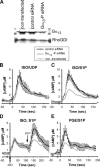
Regulation of intracellular cyclic adenosine 3 ',5 '-monophosphate (cAMP) is integral in mediating cell growth, cell differentiation, and immune responses in hematopoietic cells. To facilitate studies of cAMP regulation we developed a BRET (bioluminescence resonance energy transfer) sensor for cAMP, CAMYEL (cAMP sensor using YFP-Epac-RLuc), which can quantitatively and rapidly monitor intracellular concentrations of cAMP in vivo. This sensor was used to characterize three distinct pathways for modulation of cAMP synthesis stimulated by presumed G(s)-dependent receptors for isoproterenol and prostaglandin E(2). Whereas two ligands, uridine 5 '-diphosphate and complement C5a, appear to use known mechanisms for augmentation of cAMP via G(q)/calcium and G(i), the action of sphingosine 1-phosphate (S1P) is novel. In these cells, S1P, a biologically active lysophospholipid, greatly enhances increases in intracellular cAMP triggered by the ligands for G(s)-coupled receptors while having only a minimal effect by itself. The enhancement of cAMP by S1P is resistant to pertussis toxin and independent of intracellular calcium. Studies with RNAi and chemical perturbations demonstrate that the effect of S1P is mediated by the S1P(2) receptor and the heterotrimeric G(13) protein. Thus in these macrophage cells, all four major classes of G proteins can regulate intracellular cAMP.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

