Combining SELEX and the yeast three-hybrid system for in vivo selection and classification of RNA aptamers
- PMID: 17283213
- PMCID: PMC1831868
- DOI: 10.1261/rna.334307
Combining SELEX and the yeast three-hybrid system for in vivo selection and classification of RNA aptamers
Abstract
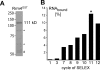
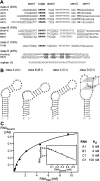
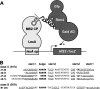
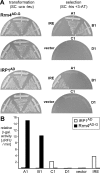
Aptamers are small nucleic acid ligands that bind to their targets with specificity and high affinity. They are generated by a combinatorial technology, known as SELEX. This in vitro approach uses iterative cycles of enrichment and amplification to select binders from nucleic acid libraries of high complexity. Here we combine SELEX with the yeast three-hybrid system in order to select for RNA aptamers with in vivo binding activity. As a target molecule, we chose the RNA recognition motif-containing RNA-binding protein Rrm4 from the corn pathogen Ustilago maydis. Rrm4 is an ELAV-like protein containing three N-terminal RNA recognition motifs (RRMs). It has been implicated in microtubule-dependent RNA transport during pathogenic development. After 11 SELEX cycles, four aptamer classes were identified. These sequences were further screened for their in vivo binding activity applying the yeast three-hybrid system. Of the initial aptamer classes only members of two classes were capable of binding in vivo. Testing representatives of both classes against Rrm4 variants mutated in one of the three RRM domains revealed that these aptamers interacted with the third RRM. Thus, the yeast three-hybrid system is a useful extension to the SELEX protocol for the identification and characterization of aptamers with in vivo binding activity.
Figures

Similar articles
-
Selection and characterization of DNA aptamers against VEGF165 with aptamer blotting method and its application.Nucleic Acids Symp Ser (Oxf). 2007;(51):399-400. doi: 10.1093/nass/nrm200. Nucleic Acids Symp Ser (Oxf). 2007. PMID: 18029755
-
Selection of RNA aptamers against recombinant transforming growth factor-beta type III receptor displayed on cell surface.Biochimie. 2006 Jul;88(7):897-904. doi: 10.1016/j.biochi.2006.02.004. Epub 2006 Mar 6. Biochimie. 2006. PMID: 16540230
-
Characterization of RNA aptamers against SRP19 protein having sequences different from SRP RNA.Nucleic Acids Symp Ser (Oxf). 2009;(53):265-6. doi: 10.1093/nass/nrp133. Nucleic Acids Symp Ser (Oxf). 2009. PMID: 19749362
-
Advances in SELEX and application of aptamers in the central nervous system.Biomol Eng. 2007 Dec;24(6):583-92. doi: 10.1016/j.bioeng.2007.06.003. Epub 2007 Jul 5. Biomol Eng. 2007. PMID: 17681489 Review.
-
SELEX--a (r)evolutionary method to generate high-affinity nucleic acid ligands.Biomol Eng. 2007 Oct;24(4):381-403. doi: 10.1016/j.bioeng.2007.06.001. Epub 2007 Jun 16. Biomol Eng. 2007. PMID: 17627883 Review.
Cited by
-
The key protein of endosomal mRNP transport Rrm4 binds translational landmark sites of cargo mRNAs.EMBO Rep. 2019 Jan;20(1):e46588. doi: 10.15252/embr.201846588. Epub 2018 Dec 14. EMBO Rep. 2019. PMID: 30552148 Free PMC article.
-
Characterization of anti-NF-kappaB RNA aptamer-binding specificity in vitro and in the yeast three-hybrid system.Nucleic Acids Res. 2009 Oct;37(18):6214-24. doi: 10.1093/nar/gkp670. Epub 2009 Aug 20. Nucleic Acids Res. 2009. PMID: 19696077 Free PMC article.
-
Tandem KH domains of Khd4 recognize AUACCC and are essential for regulation of morphology as well as pathogenicity in Ustilago maydis.RNA. 2009 Dec;15(12):2206-18. doi: 10.1261/rna.1817609. Epub 2009 Oct 23. RNA. 2009. PMID: 19854870 Free PMC article.
-
The essential roles of small non-coding RNAs and RNA modifications in normal and malignant hematopoiesis.Front Mol Biosci. 2023 Mar 31;10:1176416. doi: 10.3389/fmolb.2023.1176416. eCollection 2023. Front Mol Biosci. 2023. PMID: 37065445 Free PMC article. Review.
-
Selections that optimize RNA display in the yeast three-hybrid system.RNA. 2010 Feb;16(2):253-8. doi: 10.1261/rna.1880410. Epub 2009 Dec 14. RNA. 2010. PMID: 20008486 Free PMC article. Review.
References
-
- Becht, P., König, J., Feldbrügge, M. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J. Cell Sci. 2006;119:4964–4973. - PubMed
-
- Berezovski, M., Musheev, M., Drabovich, A., Krylov, S.N. Non-SELEX selection of aptamers. J. Am. Chem. Soc. 2006;128:1410–1411. - PubMed
-
- Bernstein, D.S., Buter, N., Stumpf, C., Wickens, M. Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods. 2002;26:123–141. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
