Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression
- PMID: 17283339
- PMCID: PMC1793903
- DOI: 10.1073/pnas.0611685104
Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression
Abstract
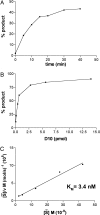
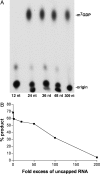
Previous studies indicated that the vaccinia virus D10 protein, which is conserved in all sequenced poxviruses, participates in the rapid turnover of host and viral mRNAs. D10 contains a motif present in the family of Nudix/MutT enzymes, a subset of which has been shown to enhance mRNA turnover in eukaryotic cells through cleavage of the 5' cap (m7GpppNm-). Here, we demonstrate that a purified recombinant D10 fusion protein possesses an intrinsic activity that liberates m7GDP from capped RNA substrates. Furthermore, point mutations in the Nudix/MutT motif abolished decapping activity. D10 has a strong affinity for capped RNA substrates (Km approximately 3 nm). RNAs of 24-309 nt were decapped to comparable extents, whereas the cap of a 12-nt RNA was uncleaved. At large molar ratios relative to capped RNA substrate, competitor m7GpppG, m7GTP, or m7GDP inhibited decapping, whereas even higher concentrations of unmethylated analogs did not. High concentrations of uncapped RNA were also inhibitory, suggesting that D10 recognizes its substrate through interaction with both cap and RNA moieties. Thus far, poxviruses represent the only virus family shown to encode a Nudix hydrolase-decapping enzyme. Although it may seem self-destructive for a virus to encode a decapping and a capping enzyme, accelerated mRNA turnover helps eliminate competing host mRNAs and allows stage-specific synthesis of viral proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses.J Virol. 2007 Dec;81(23):12973-8. doi: 10.1128/JVI.01668-07. Epub 2007 Sep 19. J Virol. 2007. PMID: 17881455 Free PMC article.
-
The Mimivirus L375 Nudix enzyme hydrolyzes the 5' mRNA cap.PLoS One. 2021 Sep 28;16(9):e0245820. doi: 10.1371/journal.pone.0245820. eCollection 2021. PLoS One. 2021. PMID: 34582446 Free PMC article.
-
A Poxvirus Decapping Enzyme Colocalizes with Mitochondria To Regulate RNA Metabolism and Translation and Promote Viral Replication.mBio. 2022 Jun 28;13(3):e0030022. doi: 10.1128/mbio.00300-22. Epub 2022 Apr 18. mBio. 2022. PMID: 35435699 Free PMC article.
-
RNA decapping inside and outside of processing bodies.Curr Opin Cell Biol. 2005 Jun;17(3):326-31. doi: 10.1016/j.ceb.2005.04.002. Curr Opin Cell Biol. 2005. PMID: 15901504 Review.
-
Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae.Annu Rev Biochem. 2000;69:571-95. doi: 10.1146/annurev.biochem.69.1.571. Annu Rev Biochem. 2000. PMID: 10966469 Review.
Cited by
-
It's the Little Things (in Viral RNA).mBio. 2020 Sep 15;11(5):e02131-20. doi: 10.1128/mBio.02131-20. mBio. 2020. PMID: 32934087 Free PMC article. Review.
-
Poxvirus-encoded decapping enzymes promote selective translation of viral mRNAs.PLoS Pathog. 2020 Oct 8;16(10):e1008926. doi: 10.1371/journal.ppat.1008926. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33031446 Free PMC article.
-
DNA-PK Is Targeted by Multiple Vaccinia Virus Proteins to Inhibit DNA Sensing.Cell Rep. 2018 Nov 13;25(7):1953-1965.e4. doi: 10.1016/j.celrep.2018.10.034. Cell Rep. 2018. PMID: 30428360 Free PMC article.
-
Characterization of the African Swine Fever Virus Decapping Enzyme during Infection.J Virol. 2017 Nov 30;91(24):e00990-17. doi: 10.1128/JVI.00990-17. Print 2017 Dec 15. J Virol. 2017. PMID: 29021398 Free PMC article.
-
Viral subversion of the host protein synthesis machinery.Nat Rev Microbiol. 2011 Oct 17;9(12):860-75. doi: 10.1038/nrmicro2655. Nat Rev Microbiol. 2011. PMID: 22002165 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

