Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe
- PMID: 17284461
- PMCID: PMC1865056
- DOI: 10.1093/nar/gkl1150
Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe
Abstract
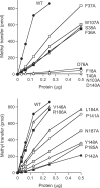
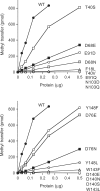
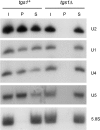
RNA cap guanine-N2 methyltransferases such as Schizosaccharomyces pombe Tgs1 and Giardia lamblia Tgs2 catalyze methylation of the exocyclic N2 amine of 7-methylguanosine. Here we performed a mutational analysis of Giardia Tgs2, entailing an alanine scan of 17 residues within the minimal active domain. Alanine substitutions at Phe18, Thr40, Asp76, Asn103 and Asp140 reduced methyltransferase specific activity to <3% of wild-type Tgs2, thereby defining these residues as essential. Alanines at Pro142, Tyr148 and Pro185 reduced activity to 7-12% of wild-type. Structure-activity relationships at Phe18, Thr40, Asp76, Asn103, Asp140 and Tyr148, and at three other essential residues defined previously (Asp68, Glu91 and Trp143) were gleaned by testing the effects of 18 conservative substitutions. Our results engender a provisional map of the Tgs2 active site, which we discuss in light of crystal structures of related methyltransferases. A genetic analysis of S. pombe Tgs1 showed that it is nonessential. An S. pombe tgs1Delta strain grows normally, notwithstanding the absence of 2,2,7-trimethylguanosine caps on its U1, U2, U4 and U5 snRNAs. However, we find that S. pombe requires cap guanine-N7 methylation catalyzed by the enzyme Pcm1. Deletion of the pcm1(+) gene was lethal, as were missense mutations in the Pcm1 active site. Thus, whereas m(7)G caps are essential in both S. pombe and S. cerevisiae, m(2,2,7)G caps are not.
Figures




Similar articles
-
RNA methyltransferases involved in 5' cap biosynthesis.RNA Biol. 2014;11(12):1597-607. doi: 10.1080/15476286.2015.1004955. RNA Biol. 2014. PMID: 25626080 Free PMC article. Review.
-
Giardia lamblia RNA cap guanine-N2 methyltransferase (Tgs2).J Biol Chem. 2005 Sep 16;280(37):32101-6. doi: 10.1074/jbc.M506438200. Epub 2005 Jul 25. J Biol Chem. 2005. PMID: 16046409
-
Cap analog substrates reveal three clades of cap guanine-N2 methyltransferases with distinct methyl acceptor specificities.RNA. 2010 Jan;16(1):211-20. doi: 10.1261/rna.1872110. Epub 2009 Nov 19. RNA. 2010. PMID: 19926722 Free PMC article.
-
Engineering Giardia lamblia trimethylguanosine synthase (GlaTgs2) to transfer non-natural modifications to the RNA 5'-cap.Protein Eng Des Sel. 2015 Jun;28(6):179-86. doi: 10.1093/protein/gzv011. Epub 2015 Mar 9. Protein Eng Des Sel. 2015. PMID: 25755274
-
The phospholipid methyltransferases in yeast.Biochim Biophys Acta. 1997 Sep 4;1348(1-2):134-41. doi: 10.1016/s0005-2760(97)00121-5. Biochim Biophys Acta. 1997. PMID: 9370325 Review.
Cited by
-
Loss of the RNA trimethylguanosine cap is compatible with nuclear accumulation of spliceosomal snRNAs but not pre-mRNA splicing or snRNA processing during animal development.PLoS Genet. 2020 Oct 21;16(10):e1009098. doi: 10.1371/journal.pgen.1009098. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33085660 Free PMC article.
-
Mutational analyses of trimethylguanosine synthase (Tgs1) and Mud2: proteins implicated in pre-mRNA splicing.RNA. 2010 May;16(5):1018-31. doi: 10.1261/rna.2082610. Epub 2010 Apr 1. RNA. 2010. PMID: 20360394 Free PMC article.
-
Genetic interactions of hypomorphic mutations in the m7G cap-binding pocket of yeast nuclear cap binding complex: an essential role for Cbc2 in meiosis via splicing of MER3 pre-mRNA.RNA. 2012 Nov;18(11):1996-2011. doi: 10.1261/rna.033746.112. Epub 2012 Sep 21. RNA. 2012. PMID: 23002122 Free PMC article.
-
Characterization of a mimivirus RNA cap guanine-N2 methyltransferase.RNA. 2009 Apr;15(4):666-74. doi: 10.1261/rna.1462109. Epub 2009 Feb 13. RNA. 2009. PMID: 19218551 Free PMC article.
-
RNA methyltransferases involved in 5' cap biosynthesis.RNA Biol. 2014;11(12):1597-607. doi: 10.1080/15476286.2015.1004955. RNA Biol. 2014. PMID: 25626080 Free PMC article. Review.
References
-
- Shuman S. The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny. Cold Spring Harbor Symp. Quant. Biol. 2001;66:301–312. - PubMed
-
- Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. mRNA capping enzyme: isolation and characterization of the gene encoding mRNA guanylyltransferase subunit from Saccharomyces cerevisiae. J. Biol. Chem. 1992;267:9521–9528. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

