Cryptic loxP sites in mammalian genomes: genome-wide distribution and relevance for the efficiency of BAC/PAC recombineering techniques
- PMID: 17284462
- PMCID: PMC1865043
- DOI: 10.1093/nar/gkl1108
Cryptic loxP sites in mammalian genomes: genome-wide distribution and relevance for the efficiency of BAC/PAC recombineering techniques
Abstract
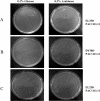
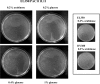
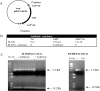
Cre is widely used for DNA tailoring and, in combination with recombineering techniques, to modify BAC/PAC sequences for generating transgenic animals. However, mammalian genomes contain recombinase recognition sites (cryptic loxP sites) that can promote illegitimate DNA recombination and damage when cells express the Cre recombinase gene. We have created a new bioinformatic tool, FuzznucComparator, which searches for cryptic loxP sites and we have applied it to the analysis of the whole mouse genome. We found that cryptic loxP sites occur frequently and are homogeneously distributed in the genome. Given the mammalian nature of BAC/PAC genomic inserts, we hypothesised that the presence of cryptic loxP sites may affect the ability to grow and modify BAC and PAC clones in E. coli expressing Cre recombinase. We have observed a defect in bacterial growth when some BACs and PACs were transformed into EL350, a DH10B-derived bacterial strain that expresses Cre recombinase under the control of an arabinose-inducible promoter. In this study, we have demonstrated that Cre recombinase expression is leaky in un-induced EL350 cells and that some BAC/PAC sequences contain cryptic loxP sites, which are active and mediate the introduction of single-strand nicks in BAC/PAC genomic inserts.
Figures




Similar articles
-
A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA.Genomics. 2001 Apr 1;73(1):56-65. doi: 10.1006/geno.2000.6451. Genomics. 2001. PMID: 11352566
-
Generation of Cre recombinase-expressing transgenic mice using bacterial artificial chromosomes.Methods Mol Biol. 2009;530:325-42. doi: 10.1007/978-1-59745-471-1_17. Methods Mol Biol. 2009. PMID: 19266340
-
Isolating large nested deletions in bacterial and P1 artificial chromosomes by in vivo P1 packaging of products of Cre-catalysed recombination between the endogenous and a transposed loxP site.Nucleic Acids Res. 1997 Jun 1;25(11):2205-12. doi: 10.1093/nar/25.11.2205. Nucleic Acids Res. 1997. PMID: 9153322 Free PMC article.
-
Cre/loxP-mediated chromosome engineering of the mouse genome.Handb Exp Pharmacol. 2007;(178):29-48. doi: 10.1007/978-3-540-35109-2_2. Handb Exp Pharmacol. 2007. PMID: 17203650 Review.
-
Recombineering: a powerful new tool for mouse functional genomics.Nat Rev Genet. 2001 Oct;2(10):769-79. doi: 10.1038/35093556. Nat Rev Genet. 2001. PMID: 11584293 Review.
Cited by
-
Mutants of Cre recombinase with improved accuracy.Nat Commun. 2013;4:2509. doi: 10.1038/ncomms3509. Nat Commun. 2013. PMID: 24056590 Free PMC article.
-
Development of mice with brain-specific deletion of floxed glud1 (glutamate dehydrogenase 1) using cre recombinase driven by the nestin promoter.Neurochem Res. 2014;39(3):456-9. doi: 10.1007/s11064-013-1041-0. Epub 2013 Apr 18. Neurochem Res. 2014. PMID: 23595828 Review.
-
CORP: Using transgenic mice to study skeletal muscle physiology.J Appl Physiol (1985). 2020 May 1;128(5):1227-1239. doi: 10.1152/japplphysiol.00021.2020. Epub 2020 Feb 27. J Appl Physiol (1985). 2020. PMID: 32105520 Free PMC article. Review.
-
ParA resolvase catalyzes site-specific excision of DNA from the Arabidopsis genome.Transgenic Res. 2009 Apr;18(2):237-48. doi: 10.1007/s11248-008-9213-4. Epub 2008 Aug 14. Transgenic Res. 2009. PMID: 18704739
-
The a"MAZE"ing world of lung-specific transgenic mice.Am J Respir Cell Mol Biol. 2012 Mar;46(3):269-82. doi: 10.1165/rcmb.2011-0372PS. Epub 2011 Dec 28. Am J Respir Cell Mol Biol. 2012. PMID: 22180870 Free PMC article. Review.
References
-
- Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. - PubMed
-
- Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. - PubMed
-
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

