The p21Waf1 pathway is involved in blocking leukemogenesis by the t(8;21) fusion protein AML1-ETO
- PMID: 17284535
- PMCID: PMC1885483
- DOI: 10.1182/blood-2006-03-012575
The p21Waf1 pathway is involved in blocking leukemogenesis by the t(8;21) fusion protein AML1-ETO
Abstract
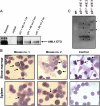
The 8;21 translocation is a major contributor to acute myeloid leukemia (AML) of the M2 classification occurring in approximately 40% of these cases. Multiple mouse models using this fusion protein demonstrate that AML1-ETO requires secondary mutagenic events to promote leukemogenesis. Here, we show that the negative cell cycle regulator p21(WAF1) gene is up-regulated by AML1-ETO at the protein, RNA, and promoter levels. Retroviral transduction and hematopoietic cell transplantation experiments with p21(WAF1)-deficient cells show that AML1-ETO is able to promote leukemogenesis in the absence of p21(WAF1). Thus, loss of p21(WAF1) facilitates AML1-ETO-induced leukemogenesis, suggesting that mutagenic events in the p21(WAF1) pathway to bypass the growth inhibitory effect from AML1-ETO-induced p21(WAF1) expression can be a significant factor in AML1-ETO-associated acute myeloid leukemia.
Figures
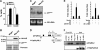


References
-
- Licht JD. AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene. 2001;20:5660–5679. - PubMed
-
- Look AT. Fusion genes and their hybrid proteins in human leukemias and lymphomas. Proc Assoc Am Physicians. 1995;107:175–180. - PubMed
-
- Mikhail FM, Sinha KK, Saunthararajah Y, Nucifora G. Normal and transforming functions of RUNX1: A perspective. J Cell Physiol. 2006;207:582–593. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

