Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini
- PMID: 17284592
- PMCID: PMC1892956
- DOI: 10.1073/pnas.0610993104
Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini
Abstract
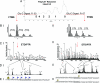
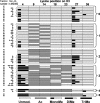
Individual posttranslational modifications (PTMs) on histones have well established roles in certain biological processes, notably transcriptional programming. Recent genomewide studies describe patterns of covalent modifications, such as H3 methylation and acetylation at promoters of specific target genes, or "bivalent domains," in stem cells, suggestive of a possible combinatorial interplay between PTMs on the same histone. However, detection of long-range PTM associations is often problematic in antibody-based or traditional mass spectrometric-based analyses. Here, histone H3 from a ciliate model was analyzed as an enriched source of transcriptionally active chromatin. Using a recently developed mass spectrometric approach, combinatorial modification states on single, long N-terminal H3 fragments (residues 1-50) were determined. The entire modification status of intact N termini was obtained and indicated correlations between K4 methylation and H3 acetylation. In addition, K4 and K27 methylation were identified concurrently on one H3 species. This methodology is applicable to other histones and larger polypeptides and will likely be a valuable tool in understanding the roles of combinatorial patterns of PTMs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- van Holde KE. Chromatin: Springer Series in Molecular Biology. New York: Springer; 1988.
-
- Wolffe AP. Chromatin: Structure and Function. San Diego: Academic; 1998.
-
- Duerre JAB, Buttz HR. In: Protein Methylation. Paik WK, Kim S, editors. Boca Raton, FL: CRC; 1990. pp. 125–138.
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. - PubMed
-
- Strahl BD, Allis CD. Nature. 2000;403:41–45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

