Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering
- PMID: 17284600
- PMCID: PMC1892959
- DOI: 10.1073/pnas.0610278104
Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering
Abstract
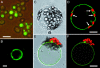
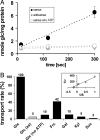
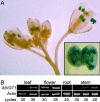
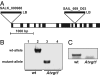
Sugar compartmentation into vacuoles of higher plants is a very important physiological process, providing extra space for transient and long-term sugar storage and contributing to the osmoregulation of cell turgor and shape. Despite the long-standing knowledge of this subcellular sugar partitioning, the proteins responsible for these transport steps have remained unknown. We have identified a gene family in Arabidopsis consisting of three members homologous to known sugar transporters. One member of this family, Arabidopsis thaliana vacuolar glucose transporter 1 (AtVGT1), was localized to the vacuolar membrane. Moreover, we provide evidence for transport activity of a tonoplast sugar transporter based on its functional expression in bakers' yeast and uptake studies in isolated yeast vacuoles. Analyses of Atvgt1 mutant lines indicate an important function of this vacuolar glucose transporter during developmental processes like seed germination and flowering.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Lemoine R. Biochim Biophys Acta. 2000;1465:246–262. - PubMed
-
- Lalonde S, Wipf D, Frommer WB. Annu Rev Plant Biol. 2004;55:341–372. - PubMed
-
- Williams LE, Lemoine R, Sauer N. Trends Plants Sci. 2000;5:283–290. - PubMed
-
- van Bel AJE. Plant Cell Environ. 2003;26:125–149.
-
- Truernit E. Curr Biol. 2001;11:R169–R171. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

