AT-rich repeats associated with chromosome 22q11.2 rearrangement disorders shape human genome architecture on Yq12
- PMID: 17284672
- PMCID: PMC1832092
- DOI: 10.1101/gr.5651507
AT-rich repeats associated with chromosome 22q11.2 rearrangement disorders shape human genome architecture on Yq12
Abstract
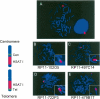
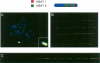
Low copy repeats (LCRs; segmental duplications) constitute approximately 5% of the sequenced human genome. Nonallelic homologous recombination events between LCRs during meiosis can lead to chromosomal rearrangements responsible for many genomic disorders. The 22q11.2 region is susceptible to recurrent and nonrecurrent deletions, duplications as well as translocations that are mediated by LCRs termed LCR22s. One particular DNA structural element, a palindromic AT-rich repeat (PATRR) present within LCR22-3a, is responsible for translocations. Similar AT-rich repeats are present within the two largest LCR22s, LCR22-2 and LCR22-4. We provide direct sequence evidence that the AT-rich repeats have altered LCR22 organization during primate evolution. The AT-rich repeats are surrounded by a subtype of human satellite I (HSAT I), and an AluSc element, forming a 2.4-kb tripartite structure. Besides 22q11.2, FISH and PCR mapping localized the tripartite repeat within heterochromatic, unsequenced regions of the genome, including the pericentromeric regions of the acrocentric chromosomes and the heterochromatic portion of Yq12 in humans. The repeat is also present on autosomes but not on chromosome Y in other hominoid species, suggesting that it has duplicated on Yq12 after speciation of humans from its common ancestor. This demonstrates that AT-rich repeats have shaped or altered the structure of the genome during evolution.
Figures

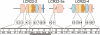


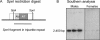

References
-
- Babcock M., Pavlicek A., Spiteri E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Pavlicek A., Spiteri E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Spiteri E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Shaffer L.G., Jurka J., Morrow B.E., Jurka J., Morrow B.E., Morrow B.E. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome Res. 2003;13:2519–2532. - PMC - PubMed
-
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Myers E.W., Li P.W., Eichler E.E., Li P.W., Eichler E.E., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. - PubMed
-
- Bandyopadhyay R., McQuillan C., Page S.L., Choo K.H., Shaffer L.G., McQuillan C., Page S.L., Choo K.H., Shaffer L.G., Page S.L., Choo K.H., Shaffer L.G., Choo K.H., Shaffer L.G., Shaffer L.G. Identification and characterization of satellite III subfamilies to the acrocentric chromosomes. Chromosome Res. 2001;9:223–233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
