Identification of muscle-specific regulatory modules in Caenorhabditis elegans
- PMID: 17284674
- PMCID: PMC1800926
- DOI: 10.1101/gr.5989907
Identification of muscle-specific regulatory modules in Caenorhabditis elegans
Abstract
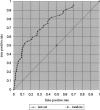
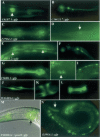
Transcriptional regulation is the major regulatory mechanism that controls the spatial and temporal expression of genes during development. This is carried out by transcription factors (TFs), which recognize and bind to their cognate binding sites. Recent studies suggest a modular organization of TF-binding sites, in which clusters of transcription-factor binding sites cooperate in the regulation of downstream gene expression. In this study, we report our computational identification and experimental verification of muscle-specific cis-regulatory modules in Caenorhabditis elegans. We first identified a set of motifs that are correlated with muscle-specific gene expression. We then predicted muscle-specific regulatory modules based on clusters of those motifs with characteristics similar to a collection of well-studied modules in other species. The method correctly identifies 88% of the experimentally characterized modules with a positive predictive value of at least 65%. The prediction accuracy of muscle-specific expression on an independent test set is highly significant (P<0.0001). We performed in vivo experimental tests of 12 predicted modules, and 10 of those drive muscle-specific gene expression. These results suggest that our method is highly accurate in identifying functional sequences important for muscle-specific gene expression and is a valuable tool for guiding experimental designs.
Figures

Similar articles
-
Conserved Motifs and Prediction of Regulatory Modules in Caenorhabditis elegans.G3 (Bethesda). 2012 Apr;2(4):469-81. doi: 10.1534/g3.111.001081. Epub 2012 Apr 1. G3 (Bethesda). 2012. PMID: 22540038 Free PMC article.
-
The Caenorhabditis elegans heterochronic regulator LIN-14 is a novel transcription factor that controls the developmental timing of transcription from the insulin/insulin-like growth factor gene ins-33 by direct DNA binding.Mol Cell Biol. 2005 Dec;25(24):11059-72. doi: 10.1128/MCB.25.24.11059-11072.2005. Mol Cell Biol. 2005. PMID: 16314527 Free PMC article.
-
The in vivo dissection of direct RFX-target gene promoters in C. elegans reveals a novel cis-regulatory element, the C-box.Dev Biol. 2012 Aug 15;368(2):415-26. doi: 10.1016/j.ydbio.2012.05.033. Epub 2012 Jun 5. Dev Biol. 2012. PMID: 22683808
-
Organizing combinatorial transcription factor recruitment at cis-regulatory modules.Transcription. 2018;9(4):233-239. doi: 10.1080/21541264.2017.1394424. Epub 2017 Nov 28. Transcription. 2018. PMID: 29105538 Free PMC article. Review.
-
Computational identification of transcriptional regulatory elements in DNA sequence.Nucleic Acids Res. 2006 Jul 19;34(12):3585-98. doi: 10.1093/nar/gkl372. Print 2006. Nucleic Acids Res. 2006. PMID: 16855295 Free PMC article. Review.
Cited by
-
TREMOR--a tool for retrieving transcriptional modules by incorporating motif covariance.Nucleic Acids Res. 2007;35(21):7360-71. doi: 10.1093/nar/gkm885. Epub 2007 Oct 25. Nucleic Acids Res. 2007. PMID: 17962303 Free PMC article.
-
Conserved elements associated with ribosomal genes and their trans-splice acceptor sites in Caenorhabditis elegans.Nucleic Acids Res. 2010 May;38(9):2990-3004. doi: 10.1093/nar/gkq003. Epub 2010 Jan 25. Nucleic Acids Res. 2010. PMID: 20100800 Free PMC article.
-
Genome-wide chromatin accessibility analyses provide a map for enhancing optic nerve regeneration.Sci Rep. 2021 Jul 21;11(1):14924. doi: 10.1038/s41598-021-94341-y. Sci Rep. 2021. PMID: 34290335 Free PMC article.
-
MORA and EnsembleTFpredictor: An ensemble approach to reveal functional transcription factor regulatory networks.PLoS One. 2023 Nov 30;18(11):e0294724. doi: 10.1371/journal.pone.0294724. eCollection 2023. PLoS One. 2023. PMID: 38032891 Free PMC article.
-
Conserved Motifs and Prediction of Regulatory Modules in Caenorhabditis elegans.G3 (Bethesda). 2012 Apr;2(4):469-81. doi: 10.1534/g3.111.001081. Epub 2012 Apr 1. G3 (Bethesda). 2012. PMID: 22540038 Free PMC article.
References
-
- Anyanful A., Sakube Y., Takuwa K., Kagawa H., Sakube Y., Takuwa K., Kagawa H., Takuwa K., Kagawa H., Kagawa H. The third and fourth tropomyosin isoforms of Caenorhabditis elegans are expressed in the pharynx and intestines and are essential for development and morphology. J. Mol. Biol. 2001;313:525–537. - PubMed
-
- Ao W., Gaudet J., Kent W.J., Muttumu S., Mango S.E., Gaudet J., Kent W.J., Muttumu S., Mango S.E., Kent W.J., Muttumu S., Mango S.E., Muttumu S., Mango S.E., Mango S.E. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science. 2004;305:1743–1746. - PubMed
-
- Arnone M.I., Davidson E.H., Davidson E.H. The hardwiring of development: Organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. - PubMed
-
- Bajic V.B., Choudhary V., Hock C.K., Choudhary V., Hock C.K., Hock C.K. Content analysis of the core promoter region of human genes. In Silico Biol. 2004;4:109–125. - PubMed
-
- Berman B.P., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Levine M., Rubin G.M., Eisen M.B., Rubin G.M., Eisen M.B., Eisen M.B. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. 2002;99:757–762. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
