Single nucleotide polymorphisms (SNPs) distinguish Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta)
- PMID: 17286860
- PMCID: PMC1803782
- DOI: 10.1186/1471-2164-8-43
Single nucleotide polymorphisms (SNPs) distinguish Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta)
Abstract
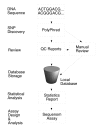
Background: Rhesus macaques serve a critical role in the study of human biomedical research. While both Indian and Chinese rhesus macaques are commonly used, genetic differences between these two subspecies affect aspects of their behavior and physiology, including response to simian immunodeficiency virus (SIV) infection. Single nucleotide polymorphisms (SNPs) can play an important role in both establishing ancestry and in identifying genes involved in complex diseases. We sequenced the 3' end of rhesus macaque genes in an effort to identify gene-based SNPs that could distinguish between Indian and Chinese rhesus macaques and aid in association analysis.
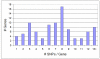
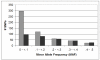
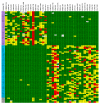
Results: We surveyed the 3' end of 94 genes in 20 rhesus macaque animals. The study included 10 animals each of Indian and Chinese ancestry. We identified a total of 661 SNPs, 457 of which appeared exclusively in one or the other population. Seventy-nine additional animals were genotyped at 44 of the population-exclusive SNPs. Of those, 38 SNPs were confirmed as being population-specific.
Conclusion: This study demonstrates that the 3' end of genes is rich in sequence polymorphisms and is suitable for the efficient discovery of gene-linked SNPs. In addition, the results show that the genomic sequences of Indian and Chinese rhesus macaque are remarkably divergent, and include numerous population-specific SNPs. These ancestral SNPs could be used for the rapid scanning of rhesus macaques, both to establish animal ancestry and to identify gene alleles that may contribute to the phenotypic differences observed in these populations.
Figures
References
-
- Kanthaswamy S, Smith DG. Effects of geographic origin on captive Macaca mulatta mitochondrial DNA variation. Comp Med. 2004;54:193–201. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

