Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates
- PMID: 17286864
- PMCID: PMC1805758
- DOI: 10.1186/1471-2164-8-45
Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates
Abstract
Background: Var genes encode a family of virulence factors known as PfEMP1 (Plasmodium falciparum erythrocyte membrane protein 1) which are responsible for both antigenic variation and cytoadherence of infected erythrocytes. Although these molecules play a central role in malaria pathogenesis, the mechanisms generating variant antigen diversification are poorly understood. To investigate var gene evolution, we compared the variant antigen repertoires from three geographically diverse parasite isolates: the 3D7 genome reference isolate; the recently sequenced HB3 isolate; and the IT4/25/5 (IT4) parasite isolate which retains the capacity to cytoadhere in vitro and in vivo.
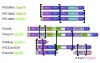
Results: These comparisons revealed that only two var genes (var1csa and var2csa) are conserved in all three isolates and one var gene (Type 3 var) has homologs in IT4 and 3D7. While the remaining 50 plus genes in each isolate are highly divergent most can be classified into the three previously defined major groups (A, B, and C) on the basis of 5' flanking sequence and chromosome location. Repertoire-wide sequence comparisons suggest that the conserved homologs are evolving separately from other var genes and that genes in group A have diverged from other groups.
Conclusion: These findings support the existence of a var gene recombination hierarchy that restricts recombination possibilities and has a central role in the functional and immunological adaptation of var genes.
Figures






References
-
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. - DOI - PubMed
-
- Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. - DOI - PubMed
-
- Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–10. doi: 10.1016/0092-8674(95)90056-X. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

