PATJ, a tight junction-associated PDZ protein, is a novel degradation target of high-risk human papillomavirus E6 and the alternatively spliced isoform 18 E6
- PMID: 17287269
- PMCID: PMC1866151
- DOI: 10.1128/JVI.02545-06
PATJ, a tight junction-associated PDZ protein, is a novel degradation target of high-risk human papillomavirus E6 and the alternatively spliced isoform 18 E6
Abstract
The E6 protein from high-risk human papillomavirus types interacts with and degrades several PDZ domain-containing proteins that localize to adherens junctions or tight junctions in polarized epithelial cells. We have identified the tight junction-associated multi-PDZ protein PATJ (PALS1-associated TJ protein) as a novel binding partner and degradation target of high-risk types 16 and 18 E6. PATJ functions in the assembly of the evolutionarily conserved CRB-PALS1-PATJ and Par6-aPKC-Par3 complexes and is critical for the formation of tight junctions in polarized cells. The ability of type 18 E6 full-length to bind to, and the subsequent degradation of, PATJ is dependent on its C-terminal PDZ binding motif. We demonstrate that the spliced 18 E6* protein, which lacks a C-terminal PDZ binding motif, associates with and degrades PATJ independently of full-length 18 E6. Thus, PATJ is the first binding partner that is degraded in response to both isoforms of 18 E6. The ability of E6 to utilize a non-E6AP ubiquitin ligase for the degradation of several PDZ binding partners has been suggested. We also demonstrate that 18 E6-mediated degradation of PATJ is not inhibited in cells where E6AP is silenced by shRNA. This suggests that the E6-E6AP complex is not required for the degradation of this protein target.
Figures


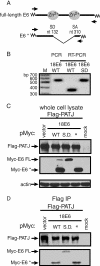




Similar articles
-
Human papillomavirus (HPV)-18 E6 oncoprotein interferes with the epithelial cell polarity Par3 protein.Mol Oncol. 2014 May;8(3):533-43. doi: 10.1016/j.molonc.2014.01.002. Epub 2014 Jan 14. Mol Oncol. 2014. PMID: 24462519 Free PMC article.
-
A systematic analysis of human papillomavirus (HPV) E6 PDZ substrates identifies MAGI-1 as a major target of HPV type 16 (HPV-16) and HPV-18 whose loss accompanies disruption of tight junctions.J Virol. 2011 Feb;85(4):1757-64. doi: 10.1128/JVI.01756-10. Epub 2010 Dec 1. J Virol. 2011. PMID: 21123374 Free PMC article.
-
Viral oncoprotein-induced mislocalization of select PDZ proteins disrupts tight junctions and causes polarity defects in epithelial cells.J Cell Sci. 2005 Sep 15;118(Pt 18):4283-93. doi: 10.1242/jcs.02560. Epub 2005 Sep 1. J Cell Sci. 2005. PMID: 16141229 Free PMC article.
-
Protein tyrosine phosphatase H1 is a target of the E6 oncoprotein of high-risk genital human papillomaviruses.J Gen Virol. 2007 Nov;88(Pt 11):2956-2965. doi: 10.1099/vir.0.83123-0. J Gen Virol. 2007. PMID: 17947517
-
Structure and function of the papillomavirus E6 protein and its interacting proteins.Front Biosci. 2008 Jan 1;13:121-34. doi: 10.2741/2664. Front Biosci. 2008. PMID: 17981532 Review.
Cited by
-
Genetic ablation of Pals1 in retinal progenitor cells models the retinal pathology of Leber congenital amaurosis.Hum Mol Genet. 2012 Jun 15;21(12):2663-76. doi: 10.1093/hmg/dds091. Epub 2012 Mar 7. Hum Mol Genet. 2012. PMID: 22398208 Free PMC article.
-
Papillomavirus E6 oncoproteins.Virology. 2013 Oct;445(1-2):115-37. doi: 10.1016/j.virol.2013.04.026. Epub 2013 May 24. Virology. 2013. PMID: 23711382 Free PMC article. Review.
-
HCV NS4B targets Scribble for proteasome-mediated degradation to facilitate cell transformation.Tumour Biol. 2016 Sep;37(9):12387-12396. doi: 10.1007/s13277-016-5100-4. Epub 2016 Jun 17. Tumour Biol. 2016. PMID: 27315218 Free PMC article.
-
The Role of E6 Spliced Isoforms (E6*) in Human Papillomavirus-Induced Carcinogenesis.Viruses. 2018 Jan 18;10(1):45. doi: 10.3390/v10010045. Viruses. 2018. PMID: 29346309 Free PMC article. Review.
-
Human papillomavirus (HPV)-18 E6 oncoprotein interferes with the epithelial cell polarity Par3 protein.Mol Oncol. 2014 May;8(3):533-43. doi: 10.1016/j.molonc.2014.01.002. Epub 2014 Jan 14. Mol Oncol. 2014. PMID: 24462519 Free PMC article.
References
-
- Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. - PubMed
-
- Doorbar, J., A. Parton, K. Hartley, L. Banks, T. Crook, M. Stanley, and L. Crawford. 1990. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology 178:254-262. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

