Motif programming: a microgene-based method for creating synthetic proteins containing multiple functional motifs
- PMID: 17287291
- PMCID: PMC1874597
- DOI: 10.1093/nar/gkm017
Motif programming: a microgene-based method for creating synthetic proteins containing multiple functional motifs
Abstract
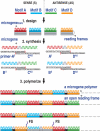
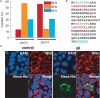
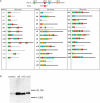
The presence of peptide motifs within the proteins provides the synthetic biologist with the opportunity to fabricate novel proteins through the programming of these motifs. Here we describe a method that enables one to combine multiple peptide motifs to generate a combinatorial protein library. With this method, a set of sense and antisense oligonucleotide primers were prepared. These primers were mixed and polymerized, so that the resultant DNA consisted of combinatorial polymers of multiple microgenes created from the stochastic assembly of the sense and antisense primers. With this motif-mixing method, we prepared a protein library from the BH1-4 motifs shared among Bcl-2 family proteins. Among the 41 clones created, 70% of clones had a stable, presumably folded expression product in human cells, which was detectable by immunohistochemistry and western blot. The proteins obtained varied with respect to both the number and the order of the four motifs. The method enables homology-independent polymerization of DNA blocks that coded motif sequences, and the frequency of each motif within a library can be adjusted in a tailor-made manner. This motif programming has a potential for creating a library with a large proportion of folded/functional proteins.
Figures



Similar articles
-
The role of peptide motifs in the evolution of a protein network.Nucleic Acids Res. 2007;35(19):6357-66. doi: 10.1093/nar/gkm692. Epub 2007 Sep 18. Nucleic Acids Res. 2007. PMID: 17881369 Free PMC article.
-
Synthesis of functional proteins by mixing peptide motifs.Chem Biol. 2004 Jun;11(6):765-73. doi: 10.1016/j.chembiol.2004.03.032. Chem Biol. 2004. PMID: 15217610
-
Natural and artificial peptide motifs: their origins and the application of motif-programming.Chem Soc Rev. 2010 Jan;39(1):117-26. doi: 10.1039/b719081f. Epub 2009 Sep 2. Chem Soc Rev. 2010. PMID: 20023842 Review.
-
[Applications of the motif-programmed proteins in medical area].Yakugaku Zasshi. 2009 Nov;129(11):1295-302. doi: 10.1248/yakushi.129.1295. Yakugaku Zasshi. 2009. PMID: 19881200 Review. Japanese.
-
Ribosome display selection of a metal-binding motif from an artificial peptide library.Biotechnol Bioeng. 2008 Dec 1;101(5):1102-7. doi: 10.1002/bit.21975. Biotechnol Bioeng. 2008. PMID: 18613123
Cited by
-
Kinetics of repeat propagation in the microgene polymerization reaction.Biophys J. 2009 Mar 4;96(5):1866-74. doi: 10.1016/j.bpj.2008.10.061. Biophys J. 2009. PMID: 19254545 Free PMC article.
-
Motif-programmed artificial protein induces apoptosis in several cancer cells by disrupting mitochondria.Cancer Sci. 2008 Feb;99(2):398-406. doi: 10.1111/j.1349-7006.2007.00697.x. Cancer Sci. 2008. PMID: 18271938 Free PMC article.
-
The role of peptide motifs in the evolution of a protein network.Nucleic Acids Res. 2007;35(19):6357-66. doi: 10.1093/nar/gkm692. Epub 2007 Sep 18. Nucleic Acids Res. 2007. PMID: 17881369 Free PMC article.
-
Synthesis of functional signaling domains by combinatorial polymerization of phosphorylation motifs.ACS Chem Biol. 2009 Sep 18;4(9):751-8. doi: 10.1021/cb900059f. ACS Chem Biol. 2009. PMID: 19627099 Free PMC article.
-
Design of an artificial phage-display library based on a new scaffold improved for average stability of the randomized proteins.Sci Rep. 2023 Jan 24;13(1):1339. doi: 10.1038/s41598-023-27710-4. Sci Rep. 2023. PMID: 36693880 Free PMC article.
References
-
- Benner SA. Synthetic biology: act natural. Nature. 2003;421:118. - PubMed
-
- Szostak JW. In vitro genetics. Trends Biochem. Sci. 1992;17:89–93. - PubMed
-
- Lee DH, Granja JR, Martinez JA, Severin K, Ghadri MR. A self-replicating peptide. Nature. 1996;382:525–528. - PubMed
-
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. - PubMed

