Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry
- PMID: 17287340
- PMCID: PMC1794346
- DOI: 10.1073/pnas.0611217104
Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry
Abstract
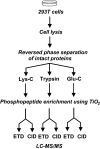
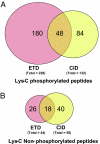
Electron transfer dissociation (ETD) is a recently introduced mass spectrometric technique that provides a more comprehensive coverage of peptide sequences and posttranslational modifications. Here, we evaluated the use of ETD for a global phosphoproteome analysis. In all, we identified a total of 1,435 phosphorylation sites from human embryonic kidney 293T cells, of which 1,141 ( approximately 80%) were not previously described. A detailed comparison of ETD and collision-induced dissociation (CID) modes showed that ETD identified 60% more phosphopeptides than CID, with an average of 40% more fragment ions that facilitated localization of phosphorylation sites. Although our data indicate that ETD is superior to CID for phosphorylation analysis, the two methods can be effectively combined in alternating ETD and CID modes for a more comprehensive analysis. Combining ETD and CID, from this single study, we were able to identify 80% of the known phosphorylation sites in >1,000 phosphorylated peptides analyzed. A hierarchical clustering of the identified phosphorylation sites allowed us to discover 15 phosphorylation motifs that have not been reported previously. Overall, ETD is an excellent method for localization of phosphorylation sites and should be an integral component of any strategy for comprehensive phosphorylation analysis.
Conflict of interest statement
Conflict of interest statement: D.M.H. and N.T. are employees of Agilent Technologies. All other authors have declared no conflict of interest.
Figures

References
-
- Haynes PA, Aebersold R. Anal Chem. 2000;72:5402–5410. - PubMed
-
- Greis KD, Hayes BK, Comer FI, Kirk M, Barnes S, Lowary TL, Hart GW. Anal Biochem. 1996;234:38–49. - PubMed
-
- Chalkley RJ, Burlingame AL. J Am Soc Mass Spectrom. 2001;12:1106–1113. - PubMed
-
- Zubarev RA, Kelleher NL, McLafferty FW. J Am Chem Soc. 1998;120:3265–3266.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

