Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry
- PMID: 17287358
- PMCID: PMC1892997
- DOI: 10.1073/pnas.0607084104
Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry
Abstract
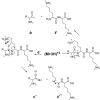
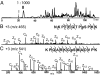
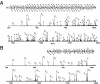
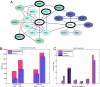
We present a strategy for the analysis of the yeast phosphoproteome that uses endo-Lys C as the proteolytic enzyme, immobilized metal affinity chromatography for phosphopeptide enrichment, a 90-min nanoflow-HPLC/electrospray-ionization MS/MS experiment for phosphopeptide fractionation and detection, gas phase ion/ion chemistry, electron transfer dissociation for peptide fragmentation, and the Open Mass Spectrometry Search Algorithm for phosphoprotein identification and assignment of phosphorylation sites. From a 30-microg (approximately 600 pmol) sample of total yeast protein, we identify 1,252 phosphorylation sites on 629 proteins. Identified phosphoproteins have expression levels that range from <50 to 1,200,000 copies per cell and are encoded by genes involved in a wide variety of cellular processes. We identify a consensus site that likely represents a motif for one or more uncharacterized kinases and show that yeast kinases, themselves, contain a disproportionately large number of phosphorylation sites. Detection of a pHis containing peptide from the yeast protein, Cdc10, suggests an unexpected role for histidine phosphorylation in septin biology. From diverse functional genomics data, we show that phosphoproteins have a higher number of interactions than an average protein and interact with each other more than with a random protein. They are also likely to be conserved across large evolutionary distances.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ficarro SB, McLeland ML, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Nat Biotechnol. 2002;20:301–305. - PubMed
-
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Nature. 2003;425:737–741. - PubMed
-
- West AH, Stock AM. Trends Biochem Sci. 2001;26:369–376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

