Ssh4, Rcr2 and Rcr1 affect plasma membrane transporter activity in Saccharomyces cerevisiae
- PMID: 17287526
- PMCID: PMC1855107
- DOI: 10.1534/genetics.106.069716
Ssh4, Rcr2 and Rcr1 affect plasma membrane transporter activity in Saccharomyces cerevisiae
Abstract
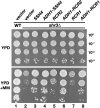
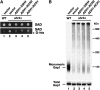
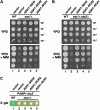
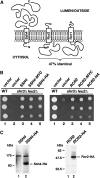
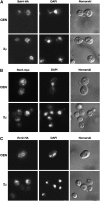
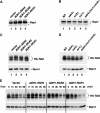
Nutrient uptake in the yeast Saccharomyces cerevisiae is a highly regulated process. Cells adjust levels of nutrient transporters within the plasma membrane at multiple stages of the secretory and endosomal pathways. In the absence of the ER-membrane-localized chaperone Shr3, amino acid permeases (AAP) inefficiently fold and are largely retained in the ER. Consequently, shr3 null mutants exhibit greatly reduced rates of amino acid uptake due to lower levels of AAPs in their plasma membranes. To further our understanding of mechanisms affecting AAP localization, we identified SSH4 and RCR2 as high-copy suppressors of shr3 null mutations. The overexpression of SSH4, RCR2, or the RCR2 homolog RCR1 increases steady-state AAP levels, whereas the genetic inactivation of these genes reduces steady-state AAP levels. Additionally, the overexpression of any of these suppressor genes exerts a positive effect on phosphate and uracil uptake systems. Ssh4 and Rcr2 primarily localize to structures associated with the vacuole; however, Rcr2 also localizes to endosome-like vesicles. Our findings are consistent with a model in which Ssh4, Rcr2, and presumably Rcr1, function within the endosome-vacuole trafficking pathway, where they affect events that determine whether plasma membrane proteins are degraded or routed to the plasma membrane.
Figures

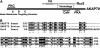
Similar articles
-
Membrane chaperone Shr3 assists in folding amino acid permeases preventing precocious ERAD.J Cell Biol. 2007 Feb 26;176(5):617-28. doi: 10.1083/jcb.200612100. J Cell Biol. 2007. PMID: 17325204 Free PMC article.
-
SHR3: a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast.Cell. 1992 Oct 30;71(3):463-78. doi: 10.1016/0092-8674(92)90515-e. Cell. 1992. PMID: 1423607
-
Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into ER-derived transport vesicles.Mol Biol Cell. 1999 Nov;10(11):3549-65. doi: 10.1091/mbc.10.11.3549. Mol Biol Cell. 1999. PMID: 10564255 Free PMC article.
-
Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae.Biochim Biophys Acta. 2005 Jul 10;1744(3):438-54. doi: 10.1016/j.bbamcr.2005.04.004. Biochim Biophys Acta. 2005. PMID: 15913810 Review.
-
The ubiquitin code of yeast permease trafficking.Trends Cell Biol. 2010 Apr;20(4):196-204. doi: 10.1016/j.tcb.2010.01.004. Trends Cell Biol. 2010. PMID: 20138522 Review.
Cited by
-
α-Arrestins and Their Functions: From Yeast to Human Health.Int J Mol Sci. 2022 Apr 30;23(9):4988. doi: 10.3390/ijms23094988. Int J Mol Sci. 2022. PMID: 35563378 Free PMC article. Review.
-
Adaptors as the regulators of HECT ubiquitin ligases.Cell Death Differ. 2021 Feb;28(2):455-472. doi: 10.1038/s41418-020-00707-6. Epub 2021 Jan 5. Cell Death Differ. 2021. PMID: 33402750 Free PMC article. Review.
-
Comparative genome-scale analysis of Pichia pastoris variants informs selection of an optimal base strain.Biotechnol Bioeng. 2020 Feb;117(2):543-555. doi: 10.1002/bit.27209. Epub 2019 Nov 12. Biotechnol Bioeng. 2020. PMID: 31654411 Free PMC article.
-
Rsp5 Ubiquitin ligase-mediated quality control system clears membrane proteins mistargeted to the vacuole membrane.J Cell Biol. 2019 Jan 7;218(1):234-250. doi: 10.1083/jcb.201806094. Epub 2018 Oct 25. J Cell Biol. 2019. PMID: 30361468 Free PMC article.
-
Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies.Mol Biol Cell. 2008 Jun;19(6):2379-88. doi: 10.1091/mbc.e08-01-0068. Epub 2008 Mar 26. Mol Biol Cell. 2008. PMID: 18367543 Free PMC article.
References
-
- Bartnicki-Garcia, S., 2006. Chitosomes: past, present and future. FEMS Yeast Res. 6: 957–965. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

