Reduced X-linked diversity in derived populations of house mice
- PMID: 17287527
- PMCID: PMC1855135
- DOI: 10.1534/genetics.106.069419
Reduced X-linked diversity in derived populations of house mice
Abstract
Contrasting patterns of X-linked vs. autosomal diversity may be indicative of the mode of selection operating in natural populations. A number of observations have shown reduced X-linked (or Z-linked) diversity relative to autosomal diversity in various organisms, suggesting a large impact of genetic hitchhiking. However, the relative contribution of other forces such as population bottlenecks, variation in reproductive success of the two sexes, and differential introgression remains unclear. Here, we survey 13 loci, 6 X-linked and 7 autosomal, in natural populations of the house mouse (Mus musculus) subspecies complex. We studied seven populations of three different subspecies, the eastern house mouse M. musculus castaneus, the central house mouse M. m. musculus, and the western house mouse M. m. domesticus, including putatively ancestral and derived populations for each. All populations display lower diversity on the X chromosomes relative to autosomes, and this effect is most pronounced in derived populations. To assess the role of demography, we fit the demographic parameters that gave the highest likelihood of the data using coalescent simulations. We find that the reduction in X-linked diversity is too large to be explained by a simple demographic model in at least two of four derived populations. These observations are also not likely to be explained by differences in reproductive success between males and females. They are consistent with a greater impact of positive selection on the X chromosome, and this is supported by the observation of an elevated K(A) and elevated K(A)/K(S) ratios on the rodent X chromosome. A second contribution may be that the X chromosome less readily introgresses across subspecies boundaries.
Figures
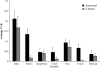
References
-
- Andolfatto, P., 2001. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18: 279–290. - PubMed
-
- Andolfatto, P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437: 1149–1152. - PubMed
-
- Aquadro, C. F., D. J. Begun and E. C. Kindahl, 1994. Selection, recombination and DNA polymorphism in Drosophila, pp. 45–56 in Non-Neutral Evolution: Theories and Molecular Data, edited by B. Golding. Chapman & Hall, New York.
-
- Avery, P. J., 1984. The population genetics of haplo-diploids and X-linked genes. Genet. Res. 44: 321–341.
-
- Bazin, E., S. Glemin and N. Galtier, 2006. Population size does not influence mitochondrial genetic diversity in animals. Science 312: 570–572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

