Assembly rules for protein networks derived from phylogenetic-statistical analysis of whole genomes
- PMID: 17288574
- PMCID: PMC1796610
- DOI: 10.1186/1471-2148-7-S1-S16
Assembly rules for protein networks derived from phylogenetic-statistical analysis of whole genomes
Abstract
Background: We report an analysis of a protein network of functionally linked proteins, identified from a phylogenetic statistical analysis of complete eukaryotic genomes. Phylogenetic methods identify pairs of proteins that co-evolve on a phylogenetic tree, and have been shown to have a high probability of correctly identifying known functional links.
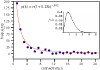
Results: The eukaryotic correlated evolution network we derive displays the familiar power law scaling of connectivity. We introduce the use of explicit phylogenetic methods to reconstruct the ancestral presence or absence of proteins at the interior nodes of a phylogeny of eukaryote species. We find that the connectivity distribution of proteins at the point they arise on the tree and join the network follows a power law, as does the connectivity distribution of proteins at the time they are lost from the network. Proteins resident in the network acquire connections over time, but we find no evidence that 'preferential attachment'--the phenomenon of newly acquired connections in the network being more likely to be made to proteins with large numbers of connections--influences the network structure. We derive a 'variable rate of attachment' model in which proteins vary in their propensity to form network interactions independently of how many connections they have or of the total number of connections in the network, and show how this model can produce apparent power-law scaling without preferential attachment.
Conclusion: A few simple rules can explain the topological structure and evolutionary changes to protein-interaction networks: most change is concentrated in satellite proteins of low connectivity and small phenotypic effect, and proteins differ in their propensity to form attachments. Given these rules of assembly, power law scaled networks naturally emerge from simple principles of selection, yielding protein interaction networks that retain a high-degree of robustness on short time scales and evolvability on longer evolutionary time scales.
Figures




References
-
- Barabasi A-Ls. Linked : the new science of networks. Cambridge, Mass.: Perseus; 2002.
-
- Albert R, Jeong H, Barabasi AL. Internet: Diameter of the World-Wide Web. Nature. 1999;401:130. doi: 10.1038/43601. - DOI
-
- Redner S. How Popular is Your Paper? An Empirical Study of the Citation Distribution. European Physical Journal B. 1998;4:131–134. doi: 10.1007/s100510050359. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

