The role of laterally transferred genes in adaptive evolution
- PMID: 17288581
- PMCID: PMC1796617
- DOI: 10.1186/1471-2148-7-S1-S8
The role of laterally transferred genes in adaptive evolution
Abstract
Background: Bacterial genomes develop new mechanisms to tide them over the imposing conditions they encounter during the course of their evolution. Acquisition of new genes by lateral gene transfer may be one of the dominant ways of adaptation in bacterial genome evolution. Lateral gene transfer provides the bacterial genome with a new set of genes that help it to explore and adapt to new ecological niches.
Methods: A maximum likelihood analysis was done on the five sequenced corynebacterial genomes to model the rates of gene insertions/deletions at various depths of the phylogeny.
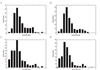
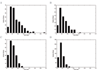
Results: The study shows that most of the laterally acquired genes are transient and the inferred rates of gene movement are higher on the external branches of the phylogeny and decrease as the phylogenetic depth increases. The newly acquired genes are under relaxed selection and evolve faster than their older counterparts. Analysis of some of the functionally characterised LGTs in each species has indicated that they may have a possible adaptive role.
Conclusion: The five Corynebacterial genomes sequenced to date have evolved by acquiring between 8-14% of their genomes by LGT and some of these genes may have a role in adaptation.
Figures


References
-
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. Massive gene decay in leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. - DOI - PubMed
-
- Foster J, Ganatra M, Kamal I, Ware J, Makarova K, Ivanova N, Bhattacharyya A, Kapatral V, Kumar S, Posfai J, Vincze T, Ingram J, Moran L, Lapidus A, Omelchenko M, Kyrpides N, Ghedin E, Wang S, Goltsman E, Joukov V, Ostrovskaya O, Tsukerman K, Mazur M, Comb D, Koonin E, Slatko B. The Wolbachia genome of Brugia malayi : endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

