"NeuroStem Chip": a novel highly specialized tool to study neural differentiation pathways in human stem cells
- PMID: 17288595
- PMCID: PMC1802744
- DOI: 10.1186/1471-2164-8-46
"NeuroStem Chip": a novel highly specialized tool to study neural differentiation pathways in human stem cells
Abstract
Background: Human stem cells are viewed as a possible source of neurons for a cell-based therapy of neurodegenerative disorders, such as Parkinson's disease. Several protocols that generate different types of neurons from human stem cells (hSCs) have been developed. Nevertheless, the cellular mechanisms that underlie the development of neurons in vitro as they are subjected to the specific differentiation protocols are often poorly understood.
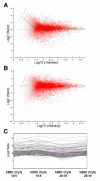
Results: We have designed a focused DNA (oligonucleotide-based) large-scale microarray platform (named "NeuroStem Chip") and used it to study gene expression patterns in hSCs as they differentiate into neurons. We have selected genes that are relevant to cells (i) being stem cells, (ii) becoming neurons, and (iii) being neurons. The NeuroStem Chip has over 1,300 pre-selected gene targets and multiple controls spotted in quadruplicates (approximately 46,000 spots total). In this study, we present the NeuroStem Chip in detail and describe the special advantages it offers to the fields of experimental neurology and stem cell biology. To illustrate the utility of NeuroStem Chip platform, we have characterized an undifferentiated population of pluripotent human embryonic stem cells (hESCs, cell line SA02). In addition, we have performed a comparative gene expression analysis of those cells versus a heterogeneous population of hESC-derived cells committed towards neuronal/dopaminergic differentiation pathway by co-culturing with PA6 stromal cells for 16 days and containing a few tyrosine hydroxylase-positive dopaminergic neurons.
Conclusion: We characterized the gene expression profiles of undifferentiated and dopaminergic lineage-committed hESC-derived cells using a highly focused custom microarray platform (NeuroStem Chip) that can become an important research tool in human stem cell biology. We propose that the areas of application for NeuroStem microarray platform could be the following: (i) characterization of the expression of established, pre-selected gene targets in hSC lines, including newly derived ones, (ii) longitudinal quality control for maintained hSC populations, (iii) following gene expression changes during differentiation under defined cell culture conditions, and (iv) confirming the success of differentiation into specific neuronal subtypes.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

