Association of six YFP-myosin XI-tail fusions with mobile plant cell organelles
- PMID: 17288617
- PMCID: PMC1802837
- DOI: 10.1186/1471-2229-7-6
Association of six YFP-myosin XI-tail fusions with mobile plant cell organelles
Abstract
Background: Myosins are molecular motors that carry cargo on actin filaments in eukaryotic cells. Seventeen myosin genes have been identified in the nuclear genome of Arabidopsis. The myosin genes can be divided into two plant-specific subfamilies, class VIII with four members and class XI with 13 members. Class XI myosins are related to animal and fungal myosin class V that are responsible for movement of particular vesicles and organelles. Organelle localization of only one of the 13 Arabidopsis myosin XI (myosin XI-6; At MYA2), which is found on peroxisomes, has so far been reported. Little information is available concerning the remaining 12 class XI myosins.
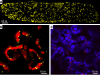
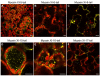
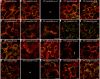
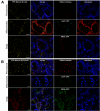
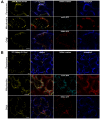
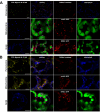
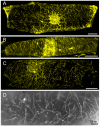
Results: We investigated 6 of the 13 class XI Arabidopsis myosins. cDNAs corresponding to the tail region of 6 myosin genes were generated and incorporated into a vector to encode YFP-myosin tail fusion proteins lacking the motor domain. Chimeric genes incorporating tail regions of myosin XI-5 (At MYA1), myosin XI-6 (At MYA2), myosin XI-8 (At XI-B), myosin XI-15 (At XI-I), myosin XI-16 (At XI-J) and myosin XI-17 (At XI-K) were expressed transiently. All YFP-myosin-tail fusion proteins were targeted to small organelles ranging in size from 0.5 to 3.0 mum. Despite the absence of a motor domain, the fluorescently-labeled organelles were motile in most cells. Tail cropping experiments demonstrated that the coiled-coil region was required for specific localization and shorter tail regions were inadequate for targeting. Myosin XI-6 (At MYA2), previously reported to localize to peroxisomes by immunofluorescence, labeled both peroxisomes and vesicles when expressed as a YFP-tail fusion. None of the 6 YFP-myosin tail fusions interacted with chloroplasts, and only one YFP-tail fusion appeared to sometimes co-localize with fluorescent proteins targeted to Golgi and mitochondria.
Conclusion: 6 myosin XI tails, extending from the coiled-coil region to the C-terminus, label specific vesicles and/or organelles when transiently expressed as YFP fusions in plant cells. Although comparable constructs lacking the motor domain result in a dominant negative effect on organelle motility in animal systems, the plant organelles remained motile. YFP-myosin tail fusions provide specific labeling for vesicles of unknown composition, whose identity can be investigated in future studies.
Figures



Similar articles
-
A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles.Plant Physiol. 2009 Jun;150(2):700-9. doi: 10.1104/pp.109.136853. Epub 2009 Apr 15. Plant Physiol. 2009. PMID: 19369591 Free PMC article.
-
Head-neck domain of Arabidopsis myosin XI, MYA2, fused with GFP produces F-actin patterns that coincide with fast organelle streaming in different plant cells.BMC Plant Biol. 2008 Jul 3;8:74. doi: 10.1186/1471-2229-8-74. BMC Plant Biol. 2008. PMID: 18598361 Free PMC article.
-
Inter-dependence of dimerization and organelle binding in myosin XI.Plant J. 2008 Aug;55(3):478-90. doi: 10.1111/j.1365-313X.2008.03522.x. Epub 2008 Apr 17. Plant J. 2008. PMID: 18429938
-
Functions of plant-specific myosin XI: from intracellular motility to plant postures.Curr Opin Plant Biol. 2015 Dec;28:30-8. doi: 10.1016/j.pbi.2015.08.006. Epub 2015 Sep 30. Curr Opin Plant Biol. 2015. PMID: 26432645 Review.
-
Actin-myosin XI: an intracellular control network in plants.Biochem Biophys Res Commun. 2018 Nov 25;506(2):403-408. doi: 10.1016/j.bbrc.2017.12.169. Epub 2018 Jan 5. Biochem Biophys Res Commun. 2018. PMID: 29307817 Review.
Cited by
-
Root hair growth: it's a one way street.F1000Prime Rep. 2015 Feb 3;7:23. doi: 10.12703/P7-23. eCollection 2015. F1000Prime Rep. 2015. PMID: 25750741 Free PMC article. Review.
-
An Update on the Role of the Actin Cytoskeleton in Plasmodesmata: A Focus on Formins.Front Plant Sci. 2021 Feb 15;12:647123. doi: 10.3389/fpls.2021.647123. eCollection 2021. Front Plant Sci. 2021. PMID: 33659020 Free PMC article. Review.
-
Light, genotype, and abscisic acid affect chloroplast positioning in guard cells of Arabidopsis thaliana leaves in distinct ways.Photosynth Res. 2010 Sep;105(3):213-27. doi: 10.1007/s11120-010-9580-6. Epub 2010 Jul 8. Photosynth Res. 2010. PMID: 20614182
-
Inhibitors of myosin, but not actin, alter transport through Tradescantia plasmodesmata.Protoplasma. 2011 Jan;248(1):205-16. doi: 10.1007/s00709-010-0244-3. Epub 2010 Nov 27. Protoplasma. 2011. PMID: 21113638
-
Rice two-pore K+ channels are expressed in different types of vacuoles.Plant Cell. 2011 Feb;23(2):756-68. doi: 10.1105/tpc.110.081463. Epub 2011 Jan 11. Plant Cell. 2011. PMID: 21224427 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

