Obscurin-like 1, OBSL1, is a novel cytoskeletal protein related to obscurin
- PMID: 17289344
- PMCID: PMC1885211
- DOI: 10.1016/j.ygeno.2006.12.004
Obscurin-like 1, OBSL1, is a novel cytoskeletal protein related to obscurin
Abstract
Cytoskeletal adaptor proteins serve vital functions in linking the internal cytoskeleton of cells to the cell membrane, particularly at sites of cell-cell and cell-matrix interactions. The importance of these adaptors to the structural integrity of the cell is evident from the number of clinical disease states attributable to defects in these networks. In the heart, defects in the cytoskeletal support system that surrounds and supports the myofibril result in dilated cardiomyopathy and congestive heart failure. In this study, we report the cloning and characterization of a novel cytoskeletal adaptor, obscurin-like 1 (OBSL1), which is closely related to obscurin, a giant structural protein required for sarcomere assembly. Multiple isoforms arise from alternative splicing, ranging in predicted molecular mass from 130 to 230 kDa. OBSL1 is located on human chromosome 2q35 within 100 kb of SPEG, another gene related to obscurin. It is expressed in a broad range of tissues and localizes to the intercalated discs, to the perinuclear region, and overlying the Z lines and M bands of adult rat cardiac myocytes. Further characterization of this novel cytoskeletal linker will have important implications for understanding the physical interactions that stabilize and support cell-matrix, cell-cell, and intracellular cytoskeletal connections.
Figures




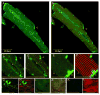

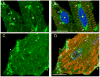
References
-
- Kostin S, Hein S, Arnon E, Scholz D, Schaper J. The cytoskeleton and related proteins in the human failing heart. Heart Fail Rev. 2000;5:271–80. - PubMed
-
- Maeda M, Holder E, Lowes B, Valent S, Bies RD. Dilated cardiomyopathy associated with deficiency of the cytoskeletal protein metavinculin. Circulation. 1997;95:17–20. - PubMed
-
- Su X, Sekiguchi M, Endo M. An ultrastructural study of cardiac myocytes in postmyocardial infarction ventricular aneurysm representative of chronic ischemic myocardium using semiquantitative and quantitative assessment. Cardiovasc Pathol. 2000;9:1–8. - PubMed
-
- Rohr S. Role of gap junctions in the propagation of the cardiac action potential. Cardiovasc Res. 2004;62:309–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

