Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex
- PMID: 17289568
- PMCID: PMC1808280
- DOI: 10.1016/j.cell.2006.12.040
Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex
Abstract
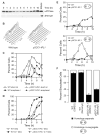
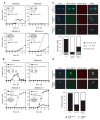
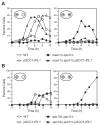
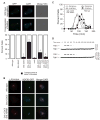
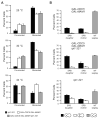
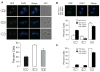
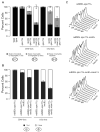
Kinetochores of sister chromatids attach to microtubules emanating from the same pole (coorientation) during meiosis I and microtubules emanating from opposite poles (biorientation) during meiosis II. We find that the Aurora B kinase Ipl1 regulates kinetochore-microtubule attachment during both meiotic divisions and that a complex known as the monopolin complex ensures that the protein kinase coorients sister chromatids during meiosis I. Furthermore, the defining of conditions sufficient to induce sister kinetochore coorientation during mitosis provides insight into monopolin complex function. The monopolin complex joins sister kinetochores independently of cohesins, the proteins that hold sister chromatids together. We propose that this function of the monopolin complex helps Aurora B coorient sister chromatids during meiosis I.
Figures
Similar articles
-
Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle.Nature. 2004 Mar 4;428(6978):93-7. doi: 10.1038/nature02328. Epub 2004 Feb 11. Nature. 2004. PMID: 14961024
-
Shugoshin promotes sister kinetochore biorientation in Saccharomyces cerevisiae.Mol Biol Cell. 2008 Mar;19(3):1199-209. doi: 10.1091/mbc.e07-06-0584. Epub 2007 Dec 19. Mol Biol Cell. 2008. PMID: 18094053 Free PMC article.
-
An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase.Dev Cell. 2003 Nov;5(5):735-45. doi: 10.1016/s1534-5807(03)00322-8. Dev Cell. 2003. PMID: 14602074
-
Structures and functions of yeast kinetochore complexes.Annu Rev Biochem. 2007;76:563-91. doi: 10.1146/annurev.biochem.76.052705.160607. Annu Rev Biochem. 2007. PMID: 17362199 Review.
-
A one-sided view of kinetochore attachment in meiosis.Cell. 2006 Sep 22;126(6):1030-2. doi: 10.1016/j.cell.2006.09.005. Cell. 2006. PMID: 16990129 Review.
Cited by
-
Meiosis in male Drosophila.Spermatogenesis. 2012 Jul 1;2(3):167-184. doi: 10.4161/spmg.21800. Spermatogenesis. 2012. PMID: 23087836 Free PMC article.
-
Temperature-dependent modulation of chromosome segregation in msh4 mutants of budding yeast.PLoS One. 2009 Oct 9;4(10):e7284. doi: 10.1371/journal.pone.0007284. PLoS One. 2009. PMID: 19816584 Free PMC article.
-
Defective histone supply causes condensin-dependent chromatin alterations, SAC activation and chromosome decatenation impairment.Nucleic Acids Res. 2014 Nov 10;42(20):12469-82. doi: 10.1093/nar/gku927. Epub 2014 Oct 9. Nucleic Acids Res. 2014. PMID: 25300489 Free PMC article.
-
Changing partners: moving from non-homologous to homologous centromere pairing in meiosis.Trends Genet. 2008 Nov;24(11):564-73. doi: 10.1016/j.tig.2008.08.006. Epub 2008 Sep 18. Trends Genet. 2008. PMID: 18804891 Free PMC article. Review.
-
Rad51 replication fork recruitment is required for DNA damage tolerance.EMBO J. 2013 May 2;32(9):1307-21. doi: 10.1038/emboj.2013.73. Epub 2013 Apr 5. EMBO J. 2013. PMID: 23563117 Free PMC article.
References
-
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. - PubMed
-
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, 3rd, Chan CS, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. - PubMed
-
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol. 2003;5:480–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

