Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors
- PMID: 17289570
- PMCID: PMC1994663
- DOI: 10.1016/j.cell.2006.12.038
Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors
Abstract
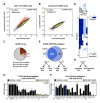
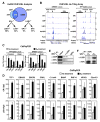
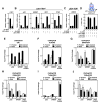
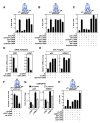
Nuclear receptors undergo ligand-dependent conformational changes that are required for corepressor-coactivator exchange, but whether there is an actual requirement for specific epigenetic landmarks to impose ligand dependency for gene activation remains unknown. Here we report an unexpected and general strategy that is based on the requirement for specific cohorts of inhibitory histone methyltransferases (HMTs) to impose gene-specific gatekeeper functions that prevent unliganded nuclear receptors and other classes of regulated transcription factors from binding to their target gene promoters and causing constitutive gene activation in the absence of stimulating signals. This strategy, based at least in part on an HMT-dependent inhibitory histone code, imposes a requirement for specific histone demethylases, including LSD1, to permit ligand- and signal-dependent activation of regulated gene expression. These events link an inhibitory methylation component of the histone code to a broadly used strategy that circumvents pathological constitutive gene induction by physiologically regulated transcription factors.
Figures

Comment in
-
Visualizing the histone code on LSD1.Cell. 2007 Feb 9;128(3):433-4. doi: 10.1016/j.cell.2007.01.017. Cell. 2007. PMID: 17289562
Similar articles
-
Visualizing the histone code on LSD1.Cell. 2007 Feb 9;128(3):433-4. doi: 10.1016/j.cell.2007.01.017. Cell. 2007. PMID: 17289562
-
LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription.Nature. 2005 Sep 15;437(7057):436-9. doi: 10.1038/nature04020. Epub 2005 Aug 3. Nature. 2005. PMID: 16079795
-
Current perspectives on histone demethylases.Acta Biochim Biophys Sin (Shanghai). 2007 Feb;39(2):81-8. doi: 10.1111/j.1745-7270.2007.00272.x. Acta Biochim Biophys Sin (Shanghai). 2007. PMID: 17277881 Review.
-
DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression.Science. 2008 Jan 11;319(5860):202-6. doi: 10.1126/science.1147674. Science. 2008. PMID: 18187655
-
[Advances in histone methyltransferases and histone demethylases].Ai Zheng. 2008 Oct;27(10):1018-25. Ai Zheng. 2008. PMID: 18851779 Review. Chinese.
Cited by
-
Proximal and distal regulation of the HYAL1 gene cluster by the estrogen receptor α in breast cancer cells.Oncotarget. 2016 Nov 22;7(47):77276-77290. doi: 10.18632/oncotarget.12630. Oncotarget. 2016. PMID: 27764788 Free PMC article.
-
Role of epigenetic modifications in luminal breast cancer.Epigenomics. 2015 Aug;7(5):847-62. doi: 10.2217/epi.15.10. Epub 2015 Feb 17. Epigenomics. 2015. PMID: 25689414 Free PMC article. Review.
-
Inducible CRISPR Epigenome Systems Mimic Cocaine Induced Bidirectional Regulation of Nab2 and Egr3.J Neurosci. 2023 Mar 29;43(13):2242-2259. doi: 10.1523/JNEUROSCI.1802-22.2022. Epub 2023 Feb 27. J Neurosci. 2023. PMID: 36849419 Free PMC article.
-
Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes.Proc Natl Acad Sci U S A. 2007 May 8;104(19):8023-8. doi: 10.1073/pnas.0700720104. Epub 2007 Apr 26. Proc Natl Acad Sci U S A. 2007. PMID: 17463086 Free PMC article.
-
Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A.Genes Dev. 2011 Mar 15;25(6):594-607. doi: 10.1101/gad.2008511. Epub 2011 Feb 28. Genes Dev. 2011. PMID: 21357675 Free PMC article.
References
-
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. - PubMed
-
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. - PubMed
-
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS034934/NS/NINDS NIH HHS/United States
- R33 CA114184/CA/NCI NIH HHS/United States
- R37 DK039949/DK/NIDDK NIH HHS/United States
- HG003119/HG/NHGRI NIH HHS/United States
- DK39949/DK/NIDDK NIH HHS/United States
- R01 CA052599/CA/NCI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- R01 CA097134/CA/NCI NIH HHS/United States
- R01 DK039949/DK/NIDDK NIH HHS/United States
- CA52599/CA/NCI NIH HHS/United States
- DK063491/DK/NIDDK NIH HHS/United States
- CA97134/CA/NCI NIH HHS/United States
- CA114184/CA/NCI NIH HHS/United States
- R01 HG003119/HG/NHGRI NIH HHS/United States
- NS34934/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

