Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex
- PMID: 17289572
- PMCID: PMC2908332
- DOI: 10.1016/j.cell.2007.01.019
Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex
Abstract
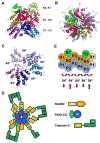
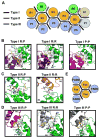
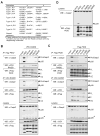
Proteins of the death domain (DD) superfamily mediate assembly of oligomeric signaling complexes for the activation of caspases and kinases via unknown mechanisms. Here we report the crystal structure of the PIDD DD and RAIDD DD complex, which forms the core of the caspase-2-activating complex PIDDosome. Although RAIDD DD and PIDD DD are monomers, they assemble into a complex that comprises seven RAIDD DDs and five PIDD DDs. Despite the use of an asymmetric assembly mechanism, all DDs in the complex are in quasi-equivalent environments. The structure provided eight unique asymmetric interfaces, which can be classified into three types. These three types of interactions together cover a majority of the DD surface. Mutagenesis on almost all interfaces leads to disruption of the assembly, resulting in defective caspase-2 activation. The three types of interactions may represent most, if not all, modes of interactions in the DD superfamily for assembling complexes of different stoichiometry.
Figures

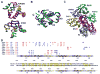

References
-
- Baliga BC, Read SH, Kumar S. The biochemical mechanism of caspase-2 activation. Cell Death Differ. 2004;11:1234–1241. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. - PubMed
-
- Duan H, Dixit VM. RAIDD is a new ‘death’ adaptor molecule. Nature. 1997;385:86–89. - PubMed
-
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

