ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP
- PMID: 17289916
- PMCID: PMC1785121
- DOI: 10.1101/gad.1521607
ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP
Abstract
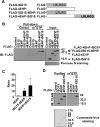
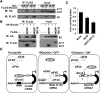
The expression of the ubiquitin-like molecule ISG15 and protein modification by ISG15 (ISGylation) are strongly activated by interferon, genotoxic stress, and pathogen infection, suggesting that ISG15 plays an important role in innate immune responses. 4EHP is an mRNA 5' cap structure-binding protein and acts as a translation suppressor by competing with eIF4E for binding to the cap structure. Here, we report that 4EHP is modified by ISG15 and ISGylated 4EHP has a much higher cap structure-binding activity. These data suggest that ISGylation of 4EHP may play an important role in cap structure-dependent translation control in immune responses.
Figures



References
-
- Cho P.F., Poulin F., Cho-Park Y.A., Cho-Park I.B., Chicoine J.D., Lasko P., Sonenberg N., Poulin F., Cho-Park Y.A., Cho-Park I.B., Chicoine J.D., Lasko P., Sonenberg N., Cho-Park Y.A., Cho-Park I.B., Chicoine J.D., Lasko P., Sonenberg N., Cho-Park I.B., Chicoine J.D., Lasko P., Sonenberg N., Chicoine J.D., Lasko P., Sonenberg N., Lasko P., Sonenberg N., Sonenberg N. A new paradigm for translational control: Inhibition via 5′–3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–423. - PubMed
-
- Dao C.T., Zhang D.E., Zhang D.E. ISG15: A ubiquitin-like enigma. Front. Biosci. 2005;10:2701–2722. - PubMed
-
- Desai S.D., Haas A.L., Wood L.M., Tsai Y.C., Pestka S., Rubin E.H., Saleem A., Nur-E-Kamal A., Liu L.F., Haas A.L., Wood L.M., Tsai Y.C., Pestka S., Rubin E.H., Saleem A., Nur-E-Kamal A., Liu L.F., Wood L.M., Tsai Y.C., Pestka S., Rubin E.H., Saleem A., Nur-E-Kamal A., Liu L.F., Tsai Y.C., Pestka S., Rubin E.H., Saleem A., Nur-E-Kamal A., Liu L.F., Pestka S., Rubin E.H., Saleem A., Nur-E-Kamal A., Liu L.F., Rubin E.H., Saleem A., Nur-E-Kamal A., Liu L.F., Saleem A., Nur-E-Kamal A., Liu L.F., Nur-E-Kamal A., Liu L.F., Liu L.F. Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res. 2006;66:921–928. - PubMed
-
- Dinkova T.D., Keiper B.D., Korneeva N.L., Aamodt E.J., Rhoads R.E., Keiper B.D., Korneeva N.L., Aamodt E.J., Rhoads R.E., Korneeva N.L., Aamodt E.J., Rhoads R.E., Aamodt E.J., Rhoads R.E., Rhoads R.E. Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol. Cell. Biol. 2005;25:100–113. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
