Death effector domain DEDa, a self-cleaved product of caspase-8/Mch5, translocates to the nucleus by binding to ERK1/2 and upregulates procaspase-8 expression via a p53-dependent mechanism
- PMID: 17290218
- PMCID: PMC1852837
- DOI: 10.1038/sj.emboj.7601571
Death effector domain DEDa, a self-cleaved product of caspase-8/Mch5, translocates to the nucleus by binding to ERK1/2 and upregulates procaspase-8 expression via a p53-dependent mechanism
Abstract
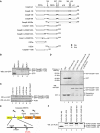
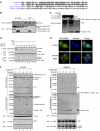
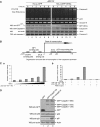
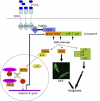
Activation of the apical caspase-8 is crucial to the extrinsic apoptotic pathway. Although the death effector domain (DED) of caspase-8 has been reported to be involved in death-inducing signaling complex formation, the detailed mechanism of how DED functions in regulating apoptosis remains largely unknown. Here, we demonstrate that the prodomain of the caspase-8/Mch5 can be further cleaved between two tandemly repeated DEDs (DEDa-DEDb) at the amino-acid residue Asp129 by caspase-8 itself. The DEDa fragment generated from the endogenous caspase-8 was detected in isolated nucleoli upon treatment with TRAIL (tumor necrosis factor-related apoptosis-inducing ligand). Cleaved DEDa appears to translocate into the nucleus by association with extracellular signal-regulated protein kinases-1/2 (ERK1/2). Elimination of ERK1/2 expression by RNA interference resulted in a significant attenuation of nuclear entry of DEDa and reduced caspase-8-dependent apoptosis. In the nucleus, DEDa interacts with TOPORS, a p53 and topoisomerase I binding protein, and possibly displaces p53 from TOPORS, allowing p53 to stimulate caspase-8 gene expression. In summary, we postulate a positive feedback loop involving DEDa, which enables the continual replenishment of procaspase-8 during apoptosis.
Figures



References
-
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI (2002) Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11 - PubMed
-
- Banelli B, Casciano I, Croce M, Di Vinci A, Gelvi I, Pagnan G, Brignole C, Allemanni G, Ferrini S, Ponzoni M, Romani M (2002) Expression and methylation of CASP8 in neuroblastoma: identification of a promoter region. Nat Med 8: 1333–1335 - PubMed
-
- Barnhart BC, Lee JC, Alappat EC, Peter ME (2003) The death effector domain protein family. Oncogene 22: 8634–8644 - PubMed
-
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS (2003) A unified model for apical caspase activation. Mol Cell 11: 529–541 - PubMed
-
- Boldin MP, Goncharov TM, Goltsev YV, Wallach D (1996) Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 85: 803–815 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

