Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin
- PMID: 17290305
- PMCID: PMC1783999
- DOI: 10.1172/JCI28523
Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin
Abstract
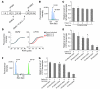
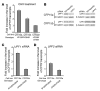
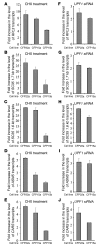
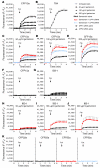
Aminoglycosides can readthrough premature termination codons (PTCs), permitting translation of full-length proteins. Previously we have found variable efficiency of readthrough in response to the aminoglycoside gentamicin among cystic fibrosis (CF) patients, all carrying the W1282X nonsense mutation. Here we demonstrate that there are patients in whom the level of CF transmembrane conductance regulator (CFTR) nonsense transcripts is markedly reduced, while in others it is significantly higher. Response to gentamicin was found only in patients with the higher level. We further investigated the possibility that the nonsense-mediated mRNA decay (NMD) might vary among cells and hence governs the level of nonsense transcripts available for readthrough. Our results demonstrate differences in NMD efficiency of CFTR transcripts carrying the W1282X mutation among different epithelial cell lines derived from the same tissue. Variability was also found for 5 physiologic NMD substrates, RPL3, SC35 1.6 kb, SC35 1.7 kb, ASNS, and CARS. Importantly, our results demonstrate the existence of cells in which NMD of all transcripts was efficient and others in which the NMD was less efficient. Downregulation of NMD in cells carrying the W1282X mutation increased the level of CFTR nonsense transcripts and enhanced the CFTR chloride channel activity in response to gentamicin. Together our results suggest that the efficiency of NMD might vary and hence have an important role in governing the response to treatments aiming to promote readthrough of PTCs in many genetic diseases.
Figures

References
-
- Frischmeyer P.A., Dietz H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999;8:1893–1900. - PubMed
-
- Martin R., Mogg A.E., Heywood L.A., Nitschke L., Burke J.F. Aminoglycoside suppression at UAG, UAA and UGA codons in Escherichia coli and human tissue culture cells. Mol. Gen. Genet. 1989;217:411–418. - PubMed
-
- Howard M., Frizzell R.A., Bedwell D.M. Aminoglycoside antibiotics restore CFTR function by overcoming premature stop mutations. Nat. Med. 1996;2:467–469. - PubMed
-
- Bedwell D.M., et al. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat. Med. 1997;3:1280–1284. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

