Mechanistic differences in DNA nanoparticle formation in the presence of oligolysines and poly-L-lysine
- PMID: 17291071
- PMCID: PMC2548297
- DOI: 10.1021/bm0605863
Mechanistic differences in DNA nanoparticle formation in the presence of oligolysines and poly-L-lysine
Abstract
We studied the effectiveness of trilysine (Lys3), tetralysine (Lys4), pentalysine (Lys5), and poly-l-lysine (PLL) (MW 50000) on lambda-DNA nanoparticle formation and characterized the size, shape, and stability of nanoparticles. Light scattering experiments showed EC50 (lysine concentration at 50% DNA compaction) values of approximately 0.0036, 2, and 20 micromol/L, respectively, for PLL, Lys5, and Lys4 at 10 mM [Na+]. Plots of log EC50 versus log [Na+] showed positive slopes of 1.09 and 1.7, respectively, for Lys4 and Lys5 and a negative slope of -0.1 for PLL. Hydrodynamic radii of oligolysine condensed particles increased (48-173 nm) with increasing [Na+], whereas no significant change occurred to nanoparticles formed with PLL. There was an increase in the size of nanoparticles formed with Lys5 at >40 degrees C, whereas no such change occurred with PLL. The DNA melting temperature increased with oligolysine concentration. These results indicate distinct differences in the mechanism(s) by which oligolysines and PLL provoke DNA condensation to nanoparticles.
Figures



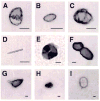

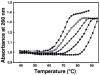

References
-
- Vijayanathan V, Thomas T, Thomas TJ. Biochemistry. 2002;41:14085. - PubMed
-
- Bloomfield VA. Curr Opin Struct Biol. 1996;6:334. - PubMed
-
- Hud NV, Vilfan ID. Annu Rev Biophys Biomol Struct. 2005;34:295. - PubMed
-
- Gosule LC, Schellman JA. J Mol Biol. 1978;121:311. - PubMed
-
- Widom J, Baldwin RL. J Mol Biol. 1980;144:431. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

