A BAC pooling strategy combined with PCR-based screenings in a large, highly repetitive genome enables integration of the maize genetic and physical maps
- PMID: 17291341
- PMCID: PMC1821331
- DOI: 10.1186/1471-2164-8-47
A BAC pooling strategy combined with PCR-based screenings in a large, highly repetitive genome enables integration of the maize genetic and physical maps
Abstract
Background: Molecular markers serve three important functions in physical map assembly. First, they provide anchor points to genetic maps facilitating functional genomic studies. Second, they reduce the overlap required for BAC contig assembly from 80 to 50 percent. Finally, they validate assemblies based solely on BAC fingerprints. We employed a six-dimensional BAC pooling strategy in combination with a high-throughput PCR-based screening method to anchor the maize genetic and physical maps.
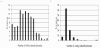
Results: A total of 110,592 maize BAC clones (approximately 6x haploid genome equivalents) were pooled into six different matrices, each containing 48 pools of BAC DNA. The quality of the BAC DNA pools and their utility for identifying BACs containing target genomic sequences was tested using 254 PCR-based STS markers. Five types of PCR-based STS markers were screened to assess potential uses for the BAC pools. An average of 4.68 BAC clones were identified per marker analyzed. These results were integrated with BAC fingerprint data generated by the Arizona Genomics Institute (AGI) and the Arizona Genomics Computational Laboratory (AGCoL) to assemble the BAC contigs using the FingerPrinted Contigs (FPC) software and contribute to the construction and anchoring of the physical map. A total of 234 markers (92.5%) anchored BAC contigs to their genetic map positions. The results can be viewed on the integrated map of maize 12.
Conclusion: This BAC pooling strategy is a rapid, cost effective method for genome assembly and anchoring. The requirement for six replicate positive amplifications makes this a robust method for use in large genomes with high amounts of repetitive DNA such as maize. This strategy can be used to physically map duplicate loci, provide order information for loci in a small genetic interval or with no genetic recombination, and loci with conflicting hybridization-based information.
Figures


References
-
- Southern EM. Application of DNA analysis to mapping the human genome. Cytogenet Cell Genet. 1982;32:52–57. - PubMed
-
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Mol Biol Rep. 1991;9:208–218.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

