Unique maturation program of the IgE response in vivo
- PMID: 17292640
- PMCID: PMC1892589
- DOI: 10.1016/j.immuni.2006.12.006
Unique maturation program of the IgE response in vivo
Abstract
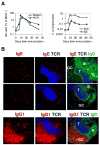
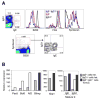
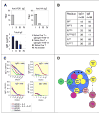
A key event in the pathogenesis of asthma and allergies is the production of IgE antibodies. We show here that IgE(+) cells were exceptional because they were largely found outside germinal centers and expressed, from very early on, a genetic program of plasma cells. In spite of their extragerminal center localization, IgE(+) cells showed signs of somatic hypermutation and affinity maturation. We demonstrated that high-affinity IgE(+) cells could be generated through a unique differentiation program that involved two phases: a pre-IgE phase in which somatic hypermutation and affinity maturation take place in IgG1(+) cells, and a post-IgE-switching phase in which IgE cells differentiate swiftly into plasma cells. Our results have implications for the understanding of IgE memory responses in allergy.
Figures




References
-
- Achatz G, Nitschke L, Lamers MC. Effect of transmembrane and cytoplasmic domains of IgE on the IgE response. Science. 1997;276:409–411. - PubMed
-
- Allen CD, Ansel KM, Low C, Lesley R, Tamamura H, Fujii N, Cyster JG. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat Immunol. 2004;5:943–952. - PubMed
-
- Coffman RL, Lebman DA, Rothman P. Mechanism and regulation of immunoglobulin isotype switching. Adv Immunol. 1993;54:229–270. - PubMed
-
- Curotto de Lafaille MA, Lino AC, Kutchukhidze N, Lafaille JJ. CD25- T cells generate CD25+Foxp3+ regulatory T cells by peripheral expansion. J Immunol. 2004;173:7259–7268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

