Changes in gene expression profiles in response to selenium supplementation among individuals with arsenic-induced pre-malignant skin lesions
- PMID: 17293063
- PMCID: PMC1924917
- DOI: 10.1016/j.toxlet.2007.01.006
Changes in gene expression profiles in response to selenium supplementation among individuals with arsenic-induced pre-malignant skin lesions
Abstract
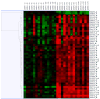
The molecular basis and downstream targets of oral selenium supplementation in individuals with elevated risk of cancer due to chronic exposure from environmental carcinogens has been largely unexplored. In this study, we investigated genome-wide differential gene expression in peripheral blood mononuclear cells (PBMC) from individuals with pre-malignant arsenic (As)-induced skin lesions before and after 6 months daily oral supplementation of 200 microg L-selenomethionine. The Affymetrix GeneChip Human 133A 2.0 array, containing probes for 22,277 gene transcripts, was used to assess gene expression. Three different normalization methods, RMA (robust multi-chip analysis), GC-RMA and PLIER (Probe logarithmic intensity error), were applied to explore differentially expressed genes. We identified a list of 28 biologically meaningful, significantly differentially expressed genes. Genes up-regulated by selenium supplementation included TNF, IL1B, IL8, SOD2, CXCL2 and several other immunological and oxidative stress-related genes. When mapped to a biological association network, many of the differentially expressed genes were found to regulate functional classes such as fibroblast growth factor, collagenase, matrix metalloproteinase and stromelysin-1, and thus, considered to affect cellular processes like apoptosis, proliferation and others. Many of the significantly up-regulated genes following selenium-supplementation were previously found by us to be down-regulated in a different set of individuals with As-induced skin lesions compared to those without. In conclusion, findings from this study may elucidate the biological effect of selenium supplementation in humans. Additionally, this study suggests that long-term selenium supplementation may revert some of the gene expression changes presumably induced by chronic As exposure in individuals with pre-malignant skin lesions.
Figures









References
-
- Affymetrix. Affymetrix GeneChip Expression analysis technical manual. Santa Clara, CA: Affymetrix; 2001.
-
- Alain G, Tousignant J, Rozenfarb E. Chronic arsenic toxicity. Int J Dermatol. 1993;32(12):899–901. - PubMed
-
- Alvaro-Gracia JM, Zvaifler NJ, Firestein GS. Cytokines in chronic inflammatory arthritis. V. Mutual antagonism between interferon-gamma and tumor necrosis factor-alpha on HLA-DR expression, proliferation, collagenase production, and granulocyte macrophage colony-stimulating factor production by rheumatoid arthritis synoviocytes. J Clin Invest. 1990;86(6):1790–1798. - PMC - PubMed
-
- Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJ., Jr Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 1999;18(24):3666–3672. - PubMed
-
- Argos M, Kibriya MG, Parvez F, Jasmine F, Rakibuz-Zaman M, Ahsan H. Gene expression profiles in peripheral lymphocytes by arsenic exposure and skin lesion status in a Bangladeshi population [accepted for publication] Cancer Epidemiol Biomarkers Prev 2006 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

