Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries
- PMID: 17293418
- PMCID: PMC1855894
- DOI: 10.1128/JB.00093-07
Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries
Abstract
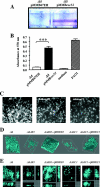
Fimbrial or nonfimbrial adhesins assembled by the bacterial chaperone-usher pathway have been demonstrated to play a key role in pathogenesis. Such an assembly mechanism has been exemplified in uropathogenic Escherichia coli strains with the Pap and the Fim systems. In Pseudomonas aeruginosa, three gene clusters (cupA, cupB, and cupC) encoding chaperone-usher pathway components have been identified in the genome sequence of the PAO1 strain. The Cup systems differ from the Pap or Fim systems, since they obviously lack numbers of genes encoding fimbrial subunits. Nevertheless, the CupA system has been demonstrated to be involved in biofilm formation on solid surfaces, whereas the role of the CupB and CupC systems in biofilm formation could not be clearly elucidated. Moreover, these gene clusters were described as poorly expressed under standard laboratory conditions. The cupB and cupC clusters are directly under the control of a two-component regulatory system designated RocA1/S1/R. In this study, we revealed that Roc1-dependent induction of the cupB and cupC genes resulted in a high level of biofilm formation, with CupB and CupC acting with synergy in clustering bacteria for microcolony formation. Very importantly, this phenotype was associated with the assembly of cell surface fimbriae visualized by electron microscopy. Finally, we observed that the CupB and CupC systems are specialized in the assembly of their own fimbrial subunits and are not exchangeable.
Figures



References
-
- Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinker, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061-1066. - PubMed
-
- de Oliveira-Garcia, D., M. Dall'Agnol, M. Rosales, A. C. Azzuz, N. Alcantara, M. B. Martinez, and J. A. Giron. 2003. Fimbriae and adherence of Stenotrophomonas maltophilia to epithelial cells and to abiotic surfaces. Cell. Microbiol. 5:625-636. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

