A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides
- PMID: 17293429
- PMCID: PMC1855832
- DOI: 10.1128/JB.01681-06
A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides
Abstract
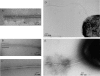
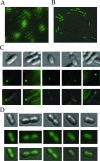
Bacteria swim in liquid environments by means of a complex rotating structure known as the flagellum. Approximately 40 proteins are required for the assembly and functionality of this structure. Rhodobacter sphaeroides has two flagellar systems. One of these systems has been shown to be functional and is required for the synthesis of the well-characterized single subpolar flagellum, while the other was found only after the genome sequence of this bacterium was completed. In this work we found that the second flagellar system of R. sphaeroides can be expressed and produces a functional flagellum. In many bacteria with two flagellar systems, one is required for swimming, while the other allows movement in denser environments by producing a large number of flagella over the entire cell surface. In contrast, the second flagellar system of R. sphaeroides produces polar flagella that are required for swimming. Expression of the second set of flagellar genes seems to be positively regulated under anaerobic growth conditions. Phylogenic analysis suggests that the flagellar system that was initially characterized was in fact acquired by horizontal transfer from a gamma-proteobacterium, while the second flagellar system contains the native genes. Interestingly, other alpha-proteobacteria closely related to R. sphaeroides have also acquired a set of flagellar genes similar to the set found in R. sphaeroides, suggesting that a common ancestor received this gene cluster.
Figures
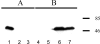



Similar articles
-
Structural Characterization of the Fla2 Flagellum of Rhodobacter sphaeroides.J Bacteriol. 2015 Sep;197(17):2859-66. doi: 10.1128/JB.00170-15. Epub 2015 Jun 29. J Bacteriol. 2015. PMID: 26124240 Free PMC article.
-
The Master Regulators of the Fla1 and Fla2 Flagella of Rhodobacter sphaeroides Control the Expression of Their Cognate CheY Proteins.J Bacteriol. 2017 Feb 14;199(5):e00670-16. doi: 10.1128/JB.00670-16. Print 2017 Mar 1. J Bacteriol. 2017. PMID: 27956523 Free PMC article.
-
Living in a Foster Home: The Single Subpolar Flagellum Fla1 of Rhodobacter sphaeroides.Biomolecules. 2020 May 16;10(5):774. doi: 10.3390/biom10050774. Biomolecules. 2020. PMID: 32429424 Free PMC article. Review.
-
A chimeric N-terminal Escherichia coli--C-terminal Rhodobacter sphaeroides FliG rotor protein supports bidirectional E. coli flagellar rotation and chemotaxis.J Bacteriol. 2005 Mar;187(5):1695-701. doi: 10.1128/JB.187.5.1695-1701.2005. J Bacteriol. 2005. PMID: 15716440 Free PMC article.
-
Dual flagellar systems enable motility under different circumstances.J Mol Microbiol Biotechnol. 2004;7(1-2):18-29. doi: 10.1159/000077866. J Mol Microbiol Biotechnol. 2004. PMID: 15170400 Review.
Cited by
-
Characterization of FlgP, an Essential Protein for Flagellar Assembly in Rhodobacter sphaeroides.J Bacteriol. 2019 Feb 11;201(5):e00752-18. doi: 10.1128/JB.00752-18. Print 2019 Mar 1. J Bacteriol. 2019. PMID: 30559113 Free PMC article.
-
Rotation of the Fla2 flagella of Cereibacter sphaeroides requires the periplasmic proteins MotK and MotE that interact with the flagellar stator protein MotB2.PLoS One. 2024 Mar 20;19(3):e0298028. doi: 10.1371/journal.pone.0298028. eCollection 2024. PLoS One. 2024. PMID: 38507361 Free PMC article.
-
Transcriptome dynamics during the transition from anaerobic photosynthesis to aerobic respiration in Rhodobacter sphaeroides 2.4.1.J Bacteriol. 2008 Jan;190(1):286-99. doi: 10.1128/JB.01375-07. Epub 2007 Oct 26. J Bacteriol. 2008. PMID: 17965166 Free PMC article.
-
Origins of flagellar gene operons and secondary flagellar systems.J Bacteriol. 2007 Oct;189(19):7098-104. doi: 10.1128/JB.00643-07. Epub 2007 Jul 20. J Bacteriol. 2007. PMID: 17644605 Free PMC article.
-
Architecture of divergent flagellar promoters controlled by CtrA in Rhodobacter sphaeroides.BMC Microbiol. 2018 Oct 10;18(1):129. doi: 10.1186/s12866-018-1264-y. BMC Microbiol. 2018. PMID: 30305031 Free PMC article.
References
-
- Abascal, F., R. Zardoya, and D. Posada. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104-2105. - PubMed
-
- Adler, J. 1966. Chemotaxis in bacteria. Science 153:708-716. - PubMed
-
- Baker, M. D., P. M. Wolanin, and J. B. Stock. 2006. Systems biology of bacterial chemotaxis. Curr. Opin. Microbiol. 9:187-192. - PubMed
-
- Ballado, T., A. Campos, L. Camarena, and G. Dreyfus. 1996. Flagellar genes from Rhodobacter sphaeroides are homologous to genes of the fliF operon of Salmonella typhimurium and to the type-III secretion system. Gene 170:69-72. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

