An early nodulin-like protein accumulates in the sieve element plasma membrane of Arabidopsis
- PMID: 17293437
- PMCID: PMC1851800
- DOI: 10.1104/pp.106.092296
An early nodulin-like protein accumulates in the sieve element plasma membrane of Arabidopsis
Abstract
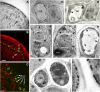
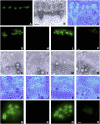
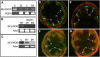
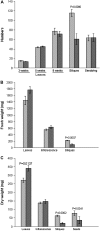
Membrane proteins within the sieve element-companion cell complex have essential roles in the physiological functioning of the phloem. The monoclonal antibody line RS6, selected from hybridomas raised against sieve elements isolated from California shield leaf (Streptanthus tortuosus; Brassicaceae) tissue cultures, recognizes an antigen in the Arabidopsis (Arabidopsis thaliana) ecotype Columbia that is associated specifically with the plasma membrane of sieve elements, but not companion cells, and accumulates at the earliest stages of sieve element differentiation. The identity of the RS6 antigen was revealed by reverse transcription-PCR of Arabidopsis leaf RNA using degenerate primers to be an early nodulin (ENOD)-like protein that is encoded by the expressed gene At3g20570. Arabidopsis ENOD-like proteins are encoded by a multigene family composed of several types of structurally related phytocyanins that have a similar overall domain structure of an amino-terminal signal peptide, plastocyanin-like copper-binding domain, proline/serine-rich domain, and carboxy-terminal hydrophobic domain. The amino- and carboxy-terminal domains of the 21.5-kD sieve element-specific ENOD are posttranslationally cleaved from the precursor protein, resulting in a mature peptide of approximately 15 kD that is attached to the sieve element plasma membrane via a carboxy-terminal glycosylphosphatidylinositol membrane anchor. Many of the Arabidopsis ENOD-like proteins accumulate in gametophytic tissues, whereas in both floral and vegetative tissues, the sieve element-specific ENOD is expressed only within the phloem. Members of the ENOD subfamily of the cupredoxin superfamily do not appear to bind copper and have unknown functions. Phenotypic analysis of homozygous T-DNA insertion mutants for the gene At3g20570 shows minimal alteration in vegetative growth but a significant reduction in the overall reproductive potential.
Figures



Similar articles
-
Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues.Plant Cell. 1999 Mar;11(3):309-22. doi: 10.1105/tpc.11.3.309. Plant Cell. 1999. PMID: 10072393 Free PMC article.
-
A plasma membrane-anchored fluorescent protein fusion illuminates sieve element plasma membranes in Arabidopsis and tobacco.Plant Physiol. 2008 Apr;146(4):1599-610. doi: 10.1104/pp.107.113274. Epub 2008 Jan 25. Plant Physiol. 2008. PMID: 18223149 Free PMC article.
-
Differential distribution of proteins expressed in companion cells in the sieve element-companion cell complex of rice plants.Plant Cell Physiol. 2005 Nov;46(11):1779-86. doi: 10.1093/pcp/pci190. Epub 2005 Aug 24. Plant Cell Physiol. 2005. PMID: 16120685
-
Phloem development.New Phytol. 2023 Aug;239(3):852-867. doi: 10.1111/nph.19003. Epub 2023 May 27. New Phytol. 2023. PMID: 37243530 Review.
-
Review: More than sweet: New insights into the biology of phloem parenchyma transfer cells in Arabidopsis.Plant Sci. 2021 Sep;310:110990. doi: 10.1016/j.plantsci.2021.110990. Epub 2021 Jul 8. Plant Sci. 2021. PMID: 34315604 Review.
Cited by
-
Transcriptome changes induced by arbuscular mycorrhizal fungi in sunflower (Helianthus annuus L.) roots.Sci Rep. 2018 Jan 8;8(1):4. doi: 10.1038/s41598-017-18445-0. Sci Rep. 2018. PMID: 29311719 Free PMC article.
-
Gene expression in mycorrhizal orchid protocorms suggests a friendly plant-fungus relationship.Planta. 2014 Jun;239(6):1337-49. doi: 10.1007/s00425-014-2062-x. Epub 2014 Apr 24. Planta. 2014. PMID: 24760407
-
Tissue-specific transcriptomic profiling of Plantago major provides insights for the involvement of vasculature in phosphate deficiency responses.Mol Genet Genomics. 2019 Feb;294(1):159-175. doi: 10.1007/s00438-018-1496-4. Epub 2018 Sep 28. Mol Genet Genomics. 2019. PMID: 30267144
-
Differential vascularization of nematode-induced feeding sites.Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12617-22. doi: 10.1073/pnas.0803835105. Epub 2008 Aug 18. Proc Natl Acad Sci U S A. 2008. PMID: 18711135 Free PMC article.
-
The putative phytocyanin genes in Chinese cabbage (Brassica rapa L.): genome-wide identification, classification and expression analysis.Mol Genet Genomics. 2013 Feb;288(1-2):1-20. doi: 10.1007/s00438-012-0726-4. Epub 2012 Dec 2. Mol Genet Genomics. 2013. PMID: 23212439
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

