The genetic code is nearly optimal for allowing additional information within protein-coding sequences
- PMID: 17293451
- PMCID: PMC1832087
- DOI: 10.1101/gr.5987307
The genetic code is nearly optimal for allowing additional information within protein-coding sequences
Abstract
DNA sequences that code for proteins need to convey, in addition to the protein-coding information, several different signals at the same time. These "parallel codes" include binding sequences for regulatory and structural proteins, signals for splicing, and RNA secondary structure. Here, we show that the universal genetic code can efficiently carry arbitrary parallel codes much better than the vast majority of other possible genetic codes. This property is related to the identity of the stop codons. We find that the ability to support parallel codes is strongly tied to another useful property of the genetic code--minimization of the effects of frame-shift translation errors. Whereas many of the known regulatory codes reside in nontranslated regions of the genome, the present findings suggest that protein-coding regions can readily carry abundant additional information.
Figures
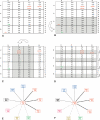
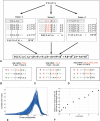
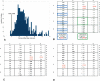

Comment in
-
Evolution and multilevel optimization of the genetic code.Genome Res. 2007 Apr;17(4):401-4. doi: 10.1101/gr.6144007. Epub 2007 Mar 9. Genome Res. 2007. PMID: 17351130 Review.
References
-
- Alon U. An introduction to systems biology. CRC Press; London, UK: 2006.
-
- Archetti M. Codon usage bias and mutation constraints reduce the level of error minimization of the genetic code. J. Mol. Evol. 2004;59:258–266. - PubMed
-
- Brooks D.J., Fresco J.R., Singh M., Fresco J.R., Singh M., Singh M. A novel method for estimating ancestral amino acid composition and its application to proteins of the Last Universal Ancestor. Bioinformatics. 2004;20:2251–2257. - PubMed
-
- Cartegni L., Chew S.L., Krainer A.R., Chew S.L., Krainer A.R., Krainer A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. - PubMed
-
- Crick F.H. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
