Characterization of the sporulation control protein SsgA by use of an efficient method to create and screen random mutant libraries in streptomycetes
- PMID: 17293502
- PMCID: PMC1855666
- DOI: 10.1128/AEM.02755-06
Characterization of the sporulation control protein SsgA by use of an efficient method to create and screen random mutant libraries in streptomycetes
Abstract
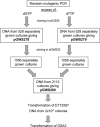
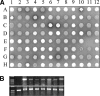
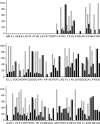
Filamentous actinomycetes are commercially widely used as producers of natural products. However, the mycelial lifestyle of actinomycetes has been a major bottleneck in their commercialization, and screening is difficult due to their poor growth on microtiter plates. We previously demonstrated that the enhanced expression of the cell division activator protein SsgA results in the fragmented growth of streptomycetes, with enhanced growth rates and improved product formation. We here describe a novel and efficient method to create, maintain, and screen mutant libraries in streptomycetes and the application of this method for the functional analysis of Streptomyces coelicolor ssgA. The variants were amplified directly from deep-frozen biomass suspensions. Around 800 ssgA variants, including single-amino-acid-substitution mutants corresponding to more than half of all SsgA residues, were analyzed for their abilities to restore sporulation to an ssgA mutant. The essential residues were clustered in three main sections, and hardly any were in the carboxy-terminal third of the protein. The majority of the crucial residues were conserved among all SsgA-like proteins (SALPs). However, the essential residues L29, D58, and S89 were conserved only in SsgA orthologues and not in other SALPs, suggesting an SsgA-specific function.
Figures

Similar articles
-
The SsgA-like proteins in actinomycetes: small proteins up to a big task.Antonie Van Leeuwenhoek. 2008 Jun;94(1):85-97. doi: 10.1007/s10482-008-9225-3. Epub 2008 Feb 14. Antonie Van Leeuwenhoek. 2008. PMID: 18273689 Free PMC article. Review.
-
A novel taxonomic marker that discriminates between morphologically complex actinomycetes.Open Biol. 2013 Oct 23;3(10):130073. doi: 10.1098/rsob.130073. Open Biol. 2013. PMID: 24153003 Free PMC article.
-
ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation.J Bacteriol. 2000 Oct;182(20):5653-62. doi: 10.1128/JB.182.20.5653-5662.2000. J Bacteriol. 2000. PMID: 11004161 Free PMC article.
-
Transcription of the sporulation gene ssgA is activated by the IclR-type regulator SsgR in a whi-independent manner in Streptomyces coelicolor A3(2).Mol Microbiol. 2004 Aug;53(3):985-1000. doi: 10.1111/j.1365-2958.2004.04186.x. Mol Microbiol. 2004. PMID: 15255907
-
Morphogenesis of Streptomyces in submerged cultures.Adv Appl Microbiol. 2014;89:1-45. doi: 10.1016/B978-0-12-800259-9.00001-9. Adv Appl Microbiol. 2014. PMID: 25131399 Review.
Cited by
-
Ectopic positioning of the cell division plane is associated with single amino acid substitutions in the FtsZ-recruiting SsgB in Streptomyces.Open Biol. 2021 Feb;11(2):200409. doi: 10.1098/rsob.200409. Epub 2021 Feb 24. Open Biol. 2021. PMID: 33622102 Free PMC article.
-
The SsgA-like proteins in actinomycetes: small proteins up to a big task.Antonie Van Leeuwenhoek. 2008 Jun;94(1):85-97. doi: 10.1007/s10482-008-9225-3. Epub 2008 Feb 14. Antonie Van Leeuwenhoek. 2008. PMID: 18273689 Free PMC article. Review.
-
A flexible mathematical model platform for studying branching networks: experimentally validated using the model actinomycete, Streptomyces coelicolor.PLoS One. 2013;8(2):e54316. doi: 10.1371/journal.pone.0054316. Epub 2013 Feb 18. PLoS One. 2013. PMID: 23441147 Free PMC article.
-
A novel taxonomic marker that discriminates between morphologically complex actinomycetes.Open Biol. 2013 Oct 23;3(10):130073. doi: 10.1098/rsob.130073. Open Biol. 2013. PMID: 24153003 Free PMC article.
References
-
- Bennett, J. W. 1998. Mycotechnology: the role of fungi in biotechnology. J. Biotechnol. 66:101-107. - PubMed
-
- Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. - PubMed
-
- Bushell, M. E. 1988. Actinomycetes in biotechnology, p. 185-217. Academic Press, London, United Kingdom.
-
- Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. - PubMed
-
- Chater, K. F. 1998. Taking a genetic scalpel to the streptomyces colony. Microbiology 144:1465-1478. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

