Cellular identification of a novel uncultured marine stramenopile (MAST-12 Clade) small-subunit rRNA gene sequence from a norwegian estuary by use of fluorescence in situ hybridization-scanning electron microscopy
- PMID: 17293516
- PMCID: PMC1855578
- DOI: 10.1128/AEM.02158-06
Cellular identification of a novel uncultured marine stramenopile (MAST-12 Clade) small-subunit rRNA gene sequence from a norwegian estuary by use of fluorescence in situ hybridization-scanning electron microscopy
Abstract
Revealing the cellular identity of organisms behind environmental eukaryote rRNA gene sequences is a major objective in microbial diversity research. We sampled an estuarine oxygen-depleted microbial mat in southwestern Norway and retrieved an 18S rRNA gene signature that branches in the MAST-12 clade, an environmental marine stramenopile clade. Detailed phylogenetic analyses revealed that MAST-12 branches among the heterotrophic stramenopiles as a sister of the free-living Bicosoecida and the parasitic genus Blastocystis. Specific sequence signatures confirmed a relationship to these two groups while excluding direct assignment. We designed a specific oligonucleotide probe for the target sequence and detected the corresponding organism in incubation samples using fluorescence in situ hybridization (FISH). Using the combined FISH-scanning electron microscopy approach (T. Stoeck, W. H. Fowle, and S. S. Epstein, Appl. Environ. Microbiol. 69:6856-6863, 2003), we determined the morphotype of the target organism among the very diverse possible morphologies of the heterotrophic stramenopiles. The unpigmented cell is spherical and about 5 mum in diameter and possesses a short flagellum and a long flagellum, both emanating anteriorly. The long flagellum bears mastigonemes in a characteristic arrangement, and its length (30 mum) distinguishes the target organism from other recognized heterotrophic stramenopiles. The short flagellum is naked and often directed posteriorly. The organism possesses neither a lorica nor a stalk. The morphological characteristics that we discovered should help isolate a representative of a novel stramenopile group, possibly at a high taxonomic level, in order to study its ultrastructure, physiological capabilities, and ecological role in the environment.
Figures
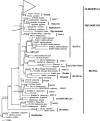



References
-
- Adl, S. M., A. G. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, S. S. Bowser, G. Brugerolle, R. A. Fensome, S. Fredericq, T. Y. James, S. Karpov, P. Kugrens, J. Krug, C. E. Lane, L. A. Lewis, J. Lodge, D. H. Lynn, D. G. Mann, R. M. McCourt, L. Mendoza, O. Moestrup, S. E. Mozley-Standridge, T. A. Nerad, C. A. Shearer, A. V. Smirnov, F. W. Spiegel, and M. F. Taylor. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399-451. - PubMed
-
- Amaral Zettler, L. A., F. Gomez, E. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Microbiology: eukaryotic diversity in Spain's River of Fire. Nature 417:137. - PubMed
-
- Baldauf, S. L., D. Bhattacharya, J. Cockrill, P. Hugenholtz, J. Pawlowski, and G. B. Simpson. 2004. The tree of life: an overview, p. 43-75. In J. Cracraft and M. J. Donoghue (ed.), Assembling the tree of life. Oxford University Press, Oxford, United Kingdom.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

