A technique for the deidentification of structural brain MR images
- PMID: 17295313
- PMCID: PMC2408762
- DOI: 10.1002/hbm.20312
A technique for the deidentification of structural brain MR images
Abstract
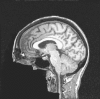
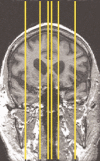
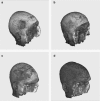
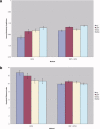
Due to the increasing need for subject privacy, the ability to deidentify structural MR images so that they do not provide full facial detail is desirable. A program was developed that uses models of nonbrain structures for removing potentially identifying facial features. When a novel image is presented, the optimal linear transform is computed for the input volume (Fischl et al. [2002]: Neuron 33:341-355; Fischl et al. [2004]: Neuroimage 23 (Suppl 1):S69-S84). A brain mask is constructed by forming the union of all voxels with nonzero probability of being brain and then morphologically dilated. All voxels outside the mask with a nonzero probability of being a facial feature are set to 0. The algorithm was applied to 342 datasets that included two different T1-weighted pulse sequences and four different diagnoses (depressed, Alzheimer's, and elderly and young control groups). Visual inspection showed none had brain tissue removed. In a detailed analysis of the impact of defacing on skull-stripping, 16 datasets were bias corrected with N3 (Sled et al. [1998]: IEEE Trans Med Imaging 17:87-97), defaced, and then skull-stripped using either a hybrid watershed algorithm (Ségonne et al. [2004]: Neuroimage 22:1060-1075, in FreeSurfer) or Brain Surface Extractor (Sandor and Leahy [1997]: IEEE Trans Med Imaging 16:41-54; Shattuck et al. [2001]: Neuroimage 13:856-876); defacing did not appreciably influence the outcome of skull-stripping. Results suggested that the automatic defacing algorithm is robust, efficiently removes nonbrain tissue, and does not unduly influence the outcome of the processing methods utilized; in some cases, skull-stripping was improved. Analyses support this algorithm as a viable method to allow data sharing with minimal data alteration within large-scale multisite projects.
Due to the increasing need for subject privacy, the ability to deidentify structural MR images so that they do not provide full facial detail is desirable. A program was developed that uses models of nonbrain structures for removing potentially identifying facial features. When a novel image is presented, the optimal linear transform is computed for the input volume (Fischl et al. [2002]: Neuron 33:341–355; Fischl et al. [2004]: Neuroimage 23 (Suppl 1):S69–S84). A brain mask is constructed by forming the union of all voxels with nonzero probability of being brain and then morphologically dilated. All voxels outside the mask with a nonzero probability of being a facial feature are set to 0. The algorithm was applied to 342 datasets that included two different T1‐weighted pulse sequences and four different diagnoses (depressed, Alzheimer's, and elderly and young control groups). Visual inspection showed none had brain tissue removed. In a detailed analysis of the impact of defacing on skull‐stripping, 16 datasets were bias corrected with N3 (Sled et al. [1998]: IEEE Trans Med Imaging 17:87–97), defaced, and then skull‐stripped using either a hybrid watershed algorithm (Ségonne et al. [2004]: Neuroimage 22:1060–1075, in FreeSurfer) or Brain Surface Extractor (Sandor and Leahy [1997]: IEEE Trans Med Imaging 16:41–54; Shattuck et al. [2001]: Neuroimage 13:856–876); defacing did not appreciably influence the outcome of skull‐stripping. Results suggested that the automatic defacing algorithm is robust, efficiently removes nonbrain tissue, and does not unduly influence the outcome of the processing methods utilized; in some cases, skull‐stripping was improved. Analyses support this algorithm as a viable method to allow data sharing with minimal data alteration within large‐scale multisite projects. Hum Brain Mapp 2007. © 2007 Wiley‐Liss, Inc.
(c) 2007 Wiley-Liss, Inc.
Figures

References
-
- Arnold JB,Liow JS,Schaper KA,Stern JJ,Sled JG,Shattuck DW,Worth AJ,Cohen MS,Leahy RM,Mazziotta JC, Rottenberg DA ( 2001): Qualitative and quantitative evaluation of six algorithms for correcting intensity nonuniformity effects. Neuroimage 13: 931–943. - PubMed
-
- Bruce V,Henderson Z,Greenwood K,Hancock PJB,Burton AM,Miller P( 1999): Verification of face identities from images captured on video. J Exp Psychol Appl 5: 339–360.
-
- Burton AM,Wilson S,Cowan M,Bruce V( 1999): Face recognition in poor‐quality video: Evidence from security surveillance. Psychol Sci 10: 243–248.
-
- Cox RW( 1996): AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29: 162–173. - PubMed
-
- Dale AM,Fischl B,Sereno MI( 1999): Cortical surface‐based analysis. I. Segmentation and surface reconstruction. Neuroimage 9: 179–194. - PubMed
Publication types
MeSH terms
Grants and funding
- P41-RR14075/RR/NCRR NIH HHS/United States
- P50 AG005131/AG/NIA NIH HHS/United States
- 5K01DA015499/DA/NIDA NIH HHS/United States
- P50 AG05131/AG/NIA NIH HHS/United States
- R01 AG004085/AG/NIA NIH HHS/United States
- 5K08MH01642/MH/NIMH NIH HHS/United States
- M01RR00827/RR/NCRR NIH HHS/United States
- R01MH42575/MH/NIMH NIH HHS/United States
- R01 RR016594/RR/NCRR NIH HHS/United States
- K08 MH001642/MH/NIMH NIH HHS/United States
- M01 RR000827/RR/NCRR NIH HHS/United States
- K01 DA015499/DA/NIDA NIH HHS/United States
- U24 RR021382/RR/NCRR NIH HHS/United States
- AG12674/AG/NIA NIH HHS/United States
- AG04085/AG/NIA NIH HHS/United States
- R01 RR16594-01A1/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

